Design and Implementation of the International Genetics and Translational Research in Transplantation Network
- PMID: 26479416
- PMCID: PMC4623847
- DOI: 10.1097/TP.0000000000000913
Design and Implementation of the International Genetics and Translational Research in Transplantation Network
Abstract
Background: Genetic association studies of transplantation outcomes have been hampered by small samples and highly complex multifactorial phenotypes, hindering investigations of the genetic architecture of a range of comorbidities which significantly impact graft and recipient life expectancy. We describe here the rationale and design of the International Genetics & Translational Research in Transplantation Network. The network comprises 22 studies to date, including 16494 transplant recipients and 11669 donors, of whom more than 5000 are of non-European ancestry, all of whom have existing genomewide genotype data sets.
Methods: We describe the rich genetic and phenotypic information available in this consortium comprising heart, kidney, liver, and lung transplant cohorts.
Results: We demonstrate significant power in International Genetics & Translational Research in Transplantation Network to detect main effect association signals across regions such as the MHC region as well as genomewide for transplant outcomes that span all solid organs, such as graft survival, acute rejection, new onset of diabetes after transplantation, and for delayed graft function in kidney only.
Conclusions: This consortium is designed and statistically powered to deliver pioneering insights into the genetic architecture of transplant-related outcomes across a range of different solid-organ transplant studies. The study design allows a spectrum of analyses to be performed including recipient-only analyses, donor-recipient HLA mismatches with focus on loss-of-function variants and nonsynonymous single nucleotide polymorphisms.
Conflict of interest statement
The authors declare no funding or conflicts of interest.
Figures
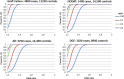
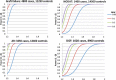
References
-
- Bamshad MJ, Ng SB, Bigham AW, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011; 12: 745– 755. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

