Increased Transcript Complexity in Genes Associated with Chronic Obstructive Pulmonary Disease
- PMID: 26480348
- PMCID: PMC4610675
- DOI: 10.1371/journal.pone.0140885
Increased Transcript Complexity in Genes Associated with Chronic Obstructive Pulmonary Disease
Abstract
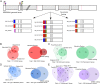
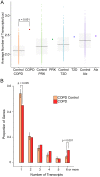
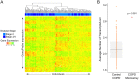
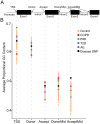
Genome-wide association studies aim to correlate genotype with phenotype. Many common diseases including Type II diabetes, Alzheimer's, Parkinson's and Chronic Obstructive Pulmonary Disease (COPD) are complex genetic traits with hundreds of different loci that are associated with varied disease risk. Identifying common features in the genes associated with each disease remains a challenge. Furthermore, the role of post-transcriptional regulation, and in particular alternative splicing, is still poorly understood in most multigenic diseases. We therefore compiled comprehensive lists of genes associated with Type II diabetes, Alzheimer's, Parkinson's and COPD in an attempt to identify common features of their corresponding mRNA transcripts within each gene set. The SERPINA1 gene is a well-recognized genetic risk factor of COPD and it produces 11 transcript variants, which is exceptional for a human gene. This led us to hypothesize that other genes associated with COPD, and complex disorders in general, are highly transcriptionally diverse. We found that COPD-associated genes have a statistically significant enrichment in transcript complexity stemming from a disproportionately high level of alternative splicing, however, Type II Diabetes, Alzheimer's and Parkinson's disease genes were not significantly enriched. We also identified a subset of transcriptionally complex COPD-associated genes (~40%) that are differentially expressed between mild, moderate and severe COPD. Although the genes associated with other lung diseases are not extensively documented, we found preliminary data that idiopathic pulmonary disease genes, but not cystic fibrosis modulators, are also more transcriptionally complex. Interestingly, complex COPD transcripts are more often the product of alternative acceptor site usage. To verify the biological importance of these alternative transcripts, we used RNA-sequencing analyses to determine that COPD-associated genes are frequently expressed in lung and liver tissues and are regulated in a tissue-specific manner. Additionally, many complex COPD-associated genes are spliced differently between COPD and non-COPD patients. Our analysis therefore suggests that post-transcriptional regulation, particularly alternative splicing, is an important feature specific to COPD disease etiology that warrants further investigation.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

