WikiPathways: capturing the full diversity of pathway knowledge
- PMID: 26481357
- PMCID: PMC4702772
- DOI: 10.1093/nar/gkv1024
WikiPathways: capturing the full diversity of pathway knowledge
Abstract
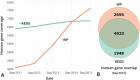
WikiPathways (http://www.wikipathways.org) is an open, collaborative platform for capturing and disseminating models of biological pathways for data visualization and analysis. Since our last NAR update, 4 years ago, WikiPathways has experienced massive growth in content, which continues to be contributed by hundreds of individuals each year. New aspects of the diversity and depth of the collected pathways are described from the perspective of researchers interested in using pathway information in their studies. We provide updates on extensions and services to support pathway analysis and visualization via popular standalone tools, i.e. PathVisio and Cytoscape, web applications and common programming environments. We introduce the Quick Edit feature for pathway authors and curators, in addition to new means of publishing pathways and maintaining custom pathway collections to serve specific research topics and communities. In addition to the latest milestones in our pathway collection and curation effort, we also highlight the latest means to access the content as publishable figures, as standard data files, and as linked data, including bulk and programmatic access.
© The Author(s) 2015. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

