Chemistry and Biology of Self-Cleaving Ribozymes
- PMID: 26481500
- PMCID: PMC4630146
- DOI: 10.1016/j.tibs.2015.09.001
Chemistry and Biology of Self-Cleaving Ribozymes
Abstract
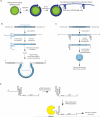
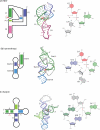
Self-cleaving ribozymes were discovered 30 years ago, but their biological distribution and catalytic mechanisms are only beginning to be defined. Each ribozyme family is defined by a distinct structure, with unique active sites accelerating the same transesterification reaction across the families. Biochemical studies show that general acid-base catalysis is the most common mechanism of self-cleavage, but metal ions and metabolites can be used as cofactors. Ribozymes have been discovered in highly diverse genomic contexts throughout nature, from viroids to vertebrates. Their biological roles include self-scission during rolling-circle replication of RNA genomes, co-transcriptional processing of retrotransposons, and metabolite-dependent gene expression regulation in bacteria. Other examples, including highly conserved mammalian ribozymes, suggest that many new biological roles are yet to be discovered.
Keywords: aptazymes; retrotransposon; riboregulation; riboswitch; ribozyme; self-scission.
Copyright © 2015 Elsevier Ltd. All rights reserved.
Figures
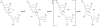

References
-
- Lai MM. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. - PubMed
-
- Taylor JM. Hepatitis delta virus. Intervirology. 1999;42:173–178. - PubMed
-
- Salehi-Ashtiani K, Luptak A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

