Conditional robustness analysis for fragility discovery and target identification in biochemical networks and in cancer systems biology
- PMID: 26482604
- PMCID: PMC4617482
- DOI: 10.1186/s12918-015-0216-5
Conditional robustness analysis for fragility discovery and target identification in biochemical networks and in cancer systems biology
Erratum in
-
Erratum to: 'Conditional robustness analysis for fragility discovery and target identification in biochemical networks and in cancer systems biology'.BMC Syst Biol. 2016 Mar 2;10:24. doi: 10.1186/s12918-016-0267-2. BMC Syst Biol. 2016. PMID: 26935321 Free PMC article. No abstract available.
Abstract
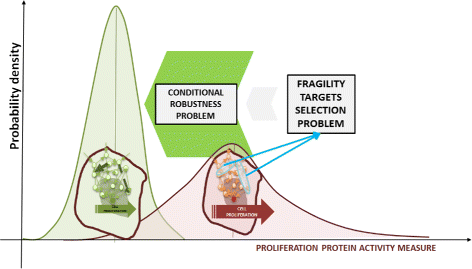
Background: The study of cancer therapy is a key issue in the field of oncology research and the development of target therapies is one of the main problems currently under investigation. This is particularly relevant in different types of tumor where traditional chemotherapy approaches often fail, such as lung cancer.
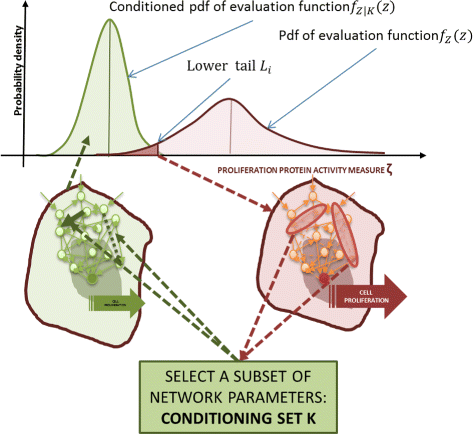
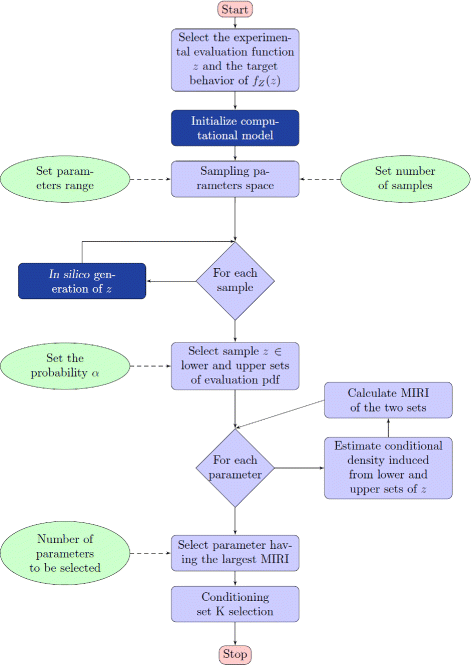
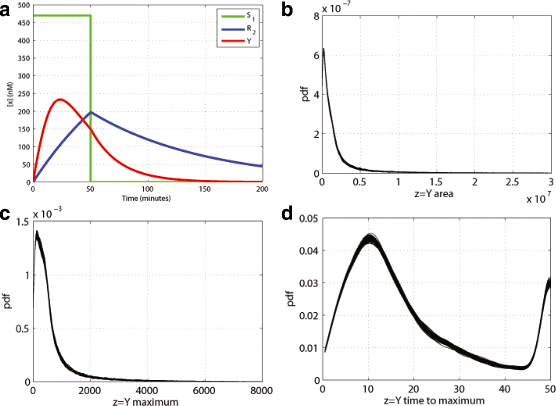
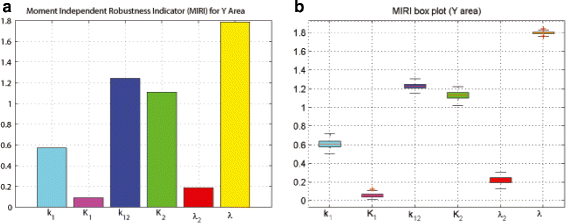
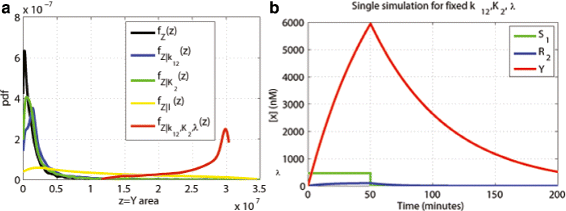
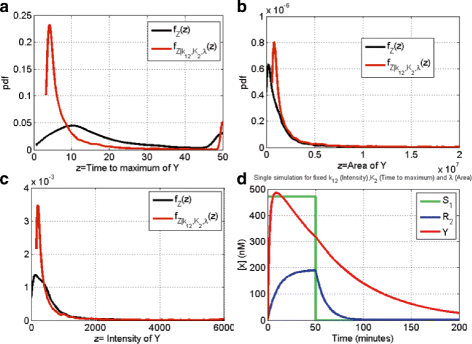
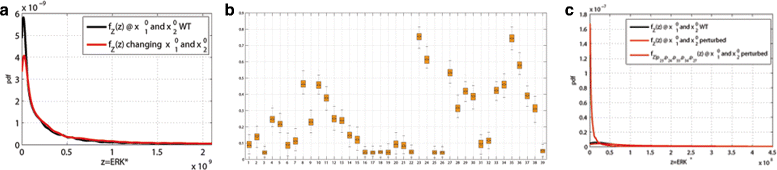
Results: We started from the general definition of robustness introduced by Kitano and applied it to the analysis of dynamical biochemical networks, proposing a new algorithm based on moment independent analysis of input/output uncertainty. The framework utilizes novel computational methods which enable evaluating the model fragility with respect to quantitative performance measures and parameters such as reaction rate constants and initial conditions. The algorithm generates a small subset of parameters that can be used to act on complex networks and to obtain the desired behaviors. We have applied the proposed framework to the EGFR-IGF1R signal transduction network, a crucial pathway in lung cancer, as an example of Cancer Systems Biology application in drug discovery. Furthermore, we have tested our framework on a pulse generator network as an example of Synthetic Biology application, thus proving the suitability of our methodology to the characterization of the input/output synthetic circuits.
Conclusions: The achieved results are of immediate practical application in computational biology, and while we demonstrate their use in two specific examples, they can in fact be used to study a wider class of biological systems.
Figures





Similar articles
-
CRA toolbox: software package for conditional robustness analysis of cancer systems biology models in MATLAB.BMC Bioinformatics. 2019 Jul 9;20(1):385. doi: 10.1186/s12859-019-2933-z. BMC Bioinformatics. 2019. PMID: 31288758 Free PMC article.
-
Gene expression complex networks: synthesis, identification, and analysis.J Comput Biol. 2011 Oct;18(10):1353-67. doi: 10.1089/cmb.2010.0118. Epub 2011 May 6. J Comput Biol. 2011. PMID: 21548810
-
A novel algorithm for finding optimal driver nodes to target control complex networks and its applications for drug targets identification.BMC Genomics. 2018 Jan 19;19(Suppl 1):924. doi: 10.1186/s12864-017-4332-z. BMC Genomics. 2018. PMID: 29363426 Free PMC article.
-
Detecting structural invariants in biological reaction networks.Methods Mol Biol. 2012;804:377-407. doi: 10.1007/978-1-61779-361-5_20. Methods Mol Biol. 2012. PMID: 22144164 Review.
-
Dynamic modelling and analysis of biochemical networks: mechanism-based models and model-based experiments.Brief Bioinform. 2006 Dec;7(4):364-74. doi: 10.1093/bib/bbl040. Epub 2006 Nov 14. Brief Bioinform. 2006. PMID: 17107967 Review.
Cited by
-
Erratum to: 'Conditional robustness analysis for fragility discovery and target identification in biochemical networks and in cancer systems biology'.BMC Syst Biol. 2016 Mar 2;10:24. doi: 10.1186/s12918-016-0267-2. BMC Syst Biol. 2016. PMID: 26935321 Free PMC article. No abstract available.
-
Application of conditional robust calibration to ordinary differential equations models in computational systems biology: a comparison of two sampling strategies.IET Syst Biol. 2020 Jun;14(3):107-119. doi: 10.1049/iet-syb.2018.5091. IET Syst Biol. 2020. PMID: 32406375 Free PMC article.
-
Reverse phase protein array (RPPA) combined with computational analysis to unravel relevant prognostic factors in non- small cell lung cancer (NSCLC): a pilot study.Oncotarget. 2017 Jun 14;8(47):83343-83353. doi: 10.18632/oncotarget.18480. eCollection 2017 Oct 10. Oncotarget. 2017. PMID: 29137348 Free PMC article.
-
Mathematical Modeling and Robustness Analysis to Unravel COVID-19 Transmission Dynamics: The Italy Case.Biology (Basel). 2020 Nov 11;9(11):394. doi: 10.3390/biology9110394. Biology (Basel). 2020. PMID: 33187109 Free PMC article.
-
CRA toolbox: software package for conditional robustness analysis of cancer systems biology models in MATLAB.BMC Bioinformatics. 2019 Jul 9;20(1):385. doi: 10.1186/s12859-019-2933-z. BMC Bioinformatics. 2019. PMID: 31288758 Free PMC article.
References
-
- Karamouzis MV, Papavassiliou AG. Tackling the cancer signal transduction “labyrinth”: A combinatorial use of biochemical tools with mathematical models will enhance the identification of optimal targets for each molecular defect. Cancer. 2014; 120(3):316–22. doi:10.1002/cncr.28424. - PubMed
-
- Wang E. Cancer systems biology. CRC Press. 2010.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

