The Nature of Genetic Variation for Complex Traits Revealed by GWAS and Regional Heritability Mapping Analyses
- PMID: 26482794
- PMCID: PMC4676519
- DOI: 10.1534/genetics.115.177220
The Nature of Genetic Variation for Complex Traits Revealed by GWAS and Regional Heritability Mapping Analyses
Abstract
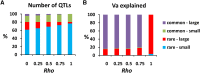
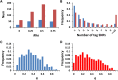
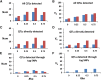
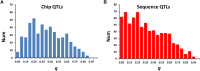
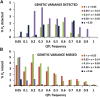
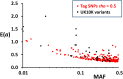
We use computer simulations to investigate the amount of genetic variation for complex traits that can be revealed by single-SNP genome-wide association studies (GWAS) or regional heritability mapping (RHM) analyses based on full genome sequence data or SNP chips. We model a large population subject to mutation, recombination, selection, and drift, assuming a pleiotropic model of mutations sampled from a bivariate distribution of effects of mutations on a quantitative trait and fitness. The pleiotropic model investigated, in contrast to previous models, implies that common mutations of large effect are responsible for most of the genetic variation for quantitative traits, except when the trait is fitness itself. We show that GWAS applied to the full sequence increases the number of QTL detected by as much as 50% compared to the number found with SNP chips but only modestly increases the amount of additive genetic variance explained. Even with full sequence data, the total amount of additive variance explained is generally below 50%. Using RHM on the full sequence data, a slightly larger number of QTL are detected than by GWAS if the same probability threshold is assumed, but these QTL explain a slightly smaller amount of genetic variance. Our results also suggest that most of the missing heritability is due to the inability to detect variants of moderate effect (∼0.03-0.3 phenotypic SDs) segregating at substantial frequencies. Very rare variants, which are more difficult to detect by GWAS, are expected to contribute little genetic variation, so their eventual detection is less relevant for resolving the missing heritability problem.
Keywords: additive genetic variance; complex traits; fitness; missing heritability; quantitative trait variation.
Copyright © 2015 by the Genetics Society of America.
Figures

References
-
- Aulchenko Y., Ripke S., Isaacs A., Van Duijn C., 2007. GenABEL: an R library for genorne-wide association analysis. Bioinformatics 23: 1294–1296. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

