Combinatorial RNA Interference Therapy Prevents Selection of Pre-existing HBV Variants in Human Liver Chimeric Mice
- PMID: 26482836
- PMCID: PMC4612501
- DOI: 10.1038/srep15259
Combinatorial RNA Interference Therapy Prevents Selection of Pre-existing HBV Variants in Human Liver Chimeric Mice
Abstract
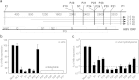
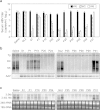
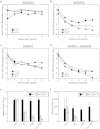
Selection of escape mutants with mutations within the target sequence could abolish the antiviral RNA interference activity. Here, we investigated the impact of a pre-existing shRNA-resistant HBV variant on the efficacy of shRNA therapy. We previously identified a highly potent shRNA, S1, which, when delivered by an adeno-associated viral vector, effectively inhibits HBV replication in HBV transgenic mice. We applied the "PICKY" software to systemically screen the HBV genome, then used hydrodynamic transfection and HBV transgenic mice to identify additional six highly potent shRNAs. Human liver chimeric mice were infected with a mixture of wild-type and T472C HBV, a S1-resistant HBV variant, and then treated with a single or combined shRNAs. The presence of T472C mutant compromised the therapeutic efficacy of S1 and resulted in replacement of serum wild-type HBV by T472C HBV. In contrast, combinatorial therapy using S1 and P28, one of six potent shRNAs, markedly reduced titers for both wild-type and T472C HBV. Interestingly, treatment with P28 alone led to the emergence of escape mutants with mutations in the P28 target region. Our results demonstrate that combinatorial RNAi therapy can minimize the escape of resistant viral mutants in chronic HBV patients.
Figures



Similar articles
-
Suppression of hepatitis B virus antigen production and replication by wild-type HBV dependently replicating HBV shRNA vectors in vitro and in vivo.Antiviral Res. 2016 Oct;134:117-129. doi: 10.1016/j.antiviral.2016.08.007. Epub 2016 Aug 30. Antiviral Res. 2016. PMID: 27591142
-
Use of RNA interference to modulate liver adenoma development in a murine model transgenic for hepatitis B virus.Gene Ther. 2012 Jan;19(1):25-33. doi: 10.1038/gt.2011.60. Epub 2011 May 12. Gene Ther. 2012. PMID: 21562593
-
Long-term inhibition of hepatitis B virus in transgenic mice by double-stranded adeno-associated virus 8-delivered short hairpin RNA.Gene Ther. 2007 Jan;14(1):11-9. doi: 10.1038/sj.gt.3302846. Epub 2006 Aug 24. Gene Ther. 2007. PMID: 16929350
-
Exploiting the RNA interference pathway to counter hepatitis B virus replication.Liver Int. 2005 Feb;25(1):9-15. doi: 10.1111/j.1478-3231.2004.0966.x. Liver Int. 2005. PMID: 15698393 Review.
-
Progress and Prospects of Anti-HBV Gene Therapy Development.Int J Mol Sci. 2015 Jul 31;16(8):17589-610. doi: 10.3390/ijms160817589. Int J Mol Sci. 2015. PMID: 26263978 Free PMC article. Review.
Cited by
-
Targeting miRNA-1a and miRNA-15b: A Novel Combinatorial Strategy to Drive Adult Cardiac Regeneration.Adv Sci (Weinh). 2025 Jun;12(21):e2414455. doi: 10.1002/advs.202414455. Epub 2025 Apr 3. Adv Sci (Weinh). 2025. PMID: 40178022 Free PMC article.
-
Gene Editing Technologies to Target HBV cccDNA.Viruses. 2022 Nov 28;14(12):2654. doi: 10.3390/v14122654. Viruses. 2022. PMID: 36560658 Free PMC article. Review.
-
RNA interference as a novel treatment strategy for chronic hepatitis B infection.Clin Mol Hepatol. 2022 Jul;28(3):408-424. doi: 10.3350/cmh.2022.0012. Epub 2022 Feb 17. Clin Mol Hepatol. 2022. PMID: 35172540 Free PMC article. Review.
-
The human C-type lectin 18 is a potential biomarker in patients with chronic hepatitis B virus infection.J Biomed Sci. 2018 Jul 28;25(1):59. doi: 10.1186/s12929-018-0460-2. J Biomed Sci. 2018. PMID: 30055605 Free PMC article.
-
Identification and characterization of proteins in the Amblyomma americanum tick cement cone.Int J Parasitol. 2018 Mar;48(3-4):211-224. doi: 10.1016/j.ijpara.2017.08.018. Epub 2017 Dec 16. Int J Parasitol. 2018. PMID: 29258831 Free PMC article.
References
-
- Iloeje U. H. et al. Predicting cirrhosis risk based on the level of circulating hepatitis B viral load. Gastroenterology 130, 678–686 (2006). - PubMed
-
- Chen C. J. et al. Risk of hepatocellular carcinoma across a biological gradient of serum hepatitis B virus DNA level. JAMA 295, 65–73 (2006). - PubMed
-
- Tseng T. C. et al. Serum hepatitis B surface antigen levels help predict disease progression in patients with low hepatitis B virus loads. Hepatology 57, 441–450 (2013). - PubMed
-
- Yang H. I. et al. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med 347, 168–174 (2002). - PubMed
-
- Scaglione S. J. & Lok A. S. Effectiveness of hepatitis B treatment in clinical practice. Gastroenterology 142, 1360–1368 e1361 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

