Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients
- PMID: 26482905
- PMCID: PMC4612410
- DOI: 10.1186/s12866-015-0563-9
Within-host microevolution of Pseudomonas aeruginosa in Italian cystic fibrosis patients
Abstract
Background: Chronic infection with Pseudomonas aeruginosa is a major cause of morbidity and mortality in cystic fibrosis (CF) patients, and a more complete understanding of P. aeruginosa within-host genomic evolution, transmission, and population genomics may provide a basis for improving intervention strategies. Here, we report the first genomic analysis of P. aeruginosa isolates sampled from Italian CF patients.
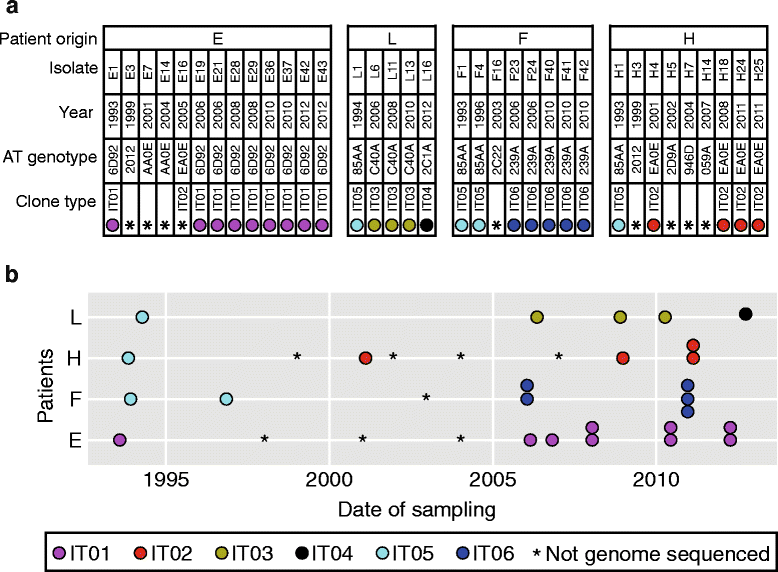
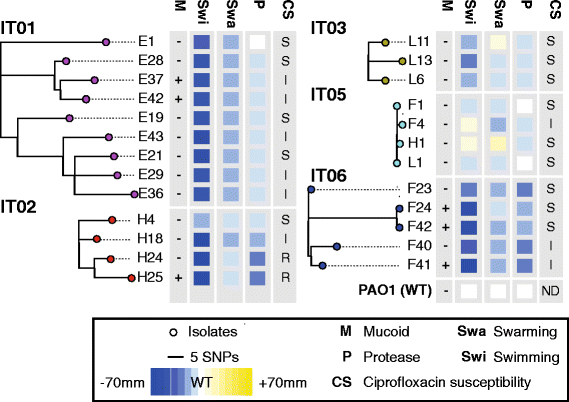
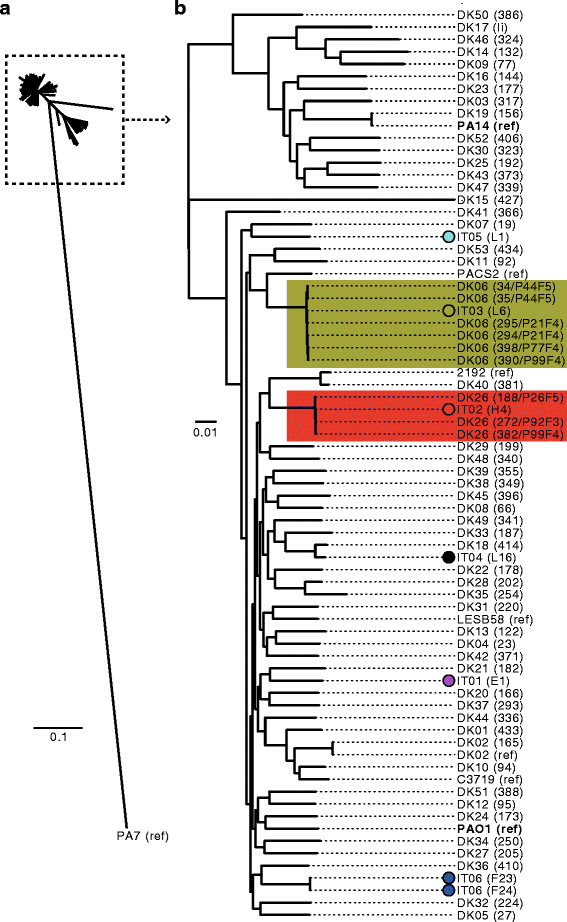
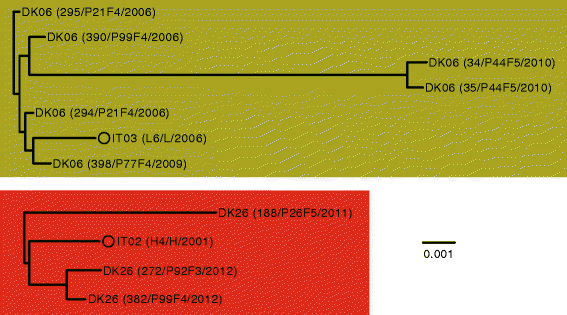
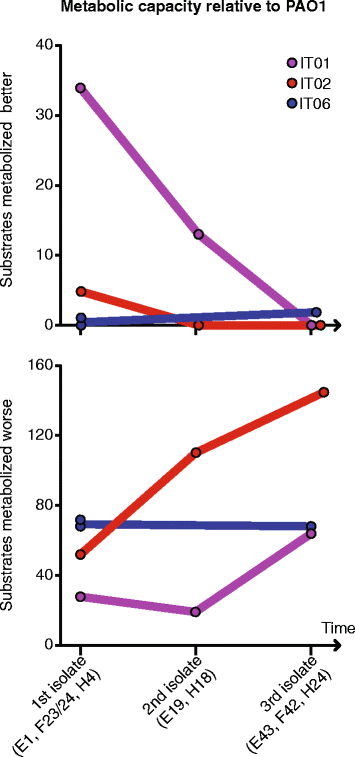
Results: By genome sequencing of 26 isolates sampled over 19 years from four patients, we elucidated the within-host evolution of clonal lineages in each individual patient. Many of the identified mutations were located in pathoadaptive genes previously associated with host adaptation, and we correlated mutations with changes in CF-relevant phenotypes such as antibiotic resistance. In addition, the genomic analysis revealed that three patients shared the same clone. Furthermore, we compared the genomes of the Italian CF isolates to a panel of genome sequenced strains of P. aeruginosa from other countries. Isolates from two of the Italian lineages belonged to clonal complexes of P. aeruginosa that have previously been identified in Danish CF patients, and our genomic comparison showed that clonal isolates from the same country may be more distantly related than clonal isolates from different countries.
Conclusions: This is the first whole-genome analysis of P. aeruginosa isolated from Italian CF patients, and together with both phenotypic and clinical information this dataset facilitates a more detailed understanding of P. aeruginosa within-host genomic evolution, transmission, and population genomics. We conclude that the evolution of the Italian lineages resembles what has been found in other countries.
Figures
Similar articles
-
Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients.Future Microbiol. 2015;10(4):599-611. doi: 10.2217/fmb.15.3. Future Microbiol. 2015. PMID: 25865196 Review.
-
Genotypic and phenotypic relatedness of Pseudomonas aeruginosa isolates among the major cystic fibrosis patient cohort in Italy.BMC Microbiol. 2016 Jul 11;16(1):142. doi: 10.1186/s12866-016-0760-1. BMC Microbiol. 2016. PMID: 27400750 Free PMC article.
-
Selective Sweeps and Parallel Pathoadaptation Drive Pseudomonas aeruginosa Evolution in the Cystic Fibrosis Lung.mBio. 2015 Sep 1;6(5):e00981-15. doi: 10.1128/mBio.00981-15. mBio. 2015. PMID: 26330513 Free PMC article.
-
Convergent evolution and adaptation of Pseudomonas aeruginosa within patients with cystic fibrosis.Nat Genet. 2015 Jan;47(1):57-64. doi: 10.1038/ng.3148. Epub 2014 Nov 17. Nat Genet. 2015. PMID: 25401299
-
Bacteriology and treatment of infections in the upper and lower airways in patients with primary ciliary dyskinesia: adressing the paranasal sinuses.Dan Med J. 2017 May;64(5):B5361. Dan Med J. 2017. PMID: 28552099 Review.
Cited by
-
Within-host evolution of bacterial pathogens during persistent infection of humans.Curr Opin Microbiol. 2022 Dec;70:102197. doi: 10.1016/j.mib.2022.102197. Epub 2022 Sep 2. Curr Opin Microbiol. 2022. PMID: 36063686 Free PMC article. Review.
-
Is genotyping of single isolates sufficient for population structure analysis of Pseudomonas aeruginosa in cystic fibrosis airways?BMC Genomics. 2016 Aug 9;17:589. doi: 10.1186/s12864-016-2873-1. BMC Genomics. 2016. PMID: 27506816 Free PMC article.
-
The Nutritional Environment Is Sufficient To Select Coexisting Biofilm and Quorum Sensing Mutants of Pseudomonas aeruginosa.J Bacteriol. 2022 Mar 15;204(3):e0044421. doi: 10.1128/JB.00444-21. Epub 2022 Jan 3. J Bacteriol. 2022. PMID: 34978461 Free PMC article.
-
Uncovering a hidden functional role of the XRE-cupin protein PsdR as a novel quorum-sensing regulator in Pseudomonas aeruginosa.PLoS Pathog. 2024 Mar 14;20(3):e1012078. doi: 10.1371/journal.ppat.1012078. eCollection 2024 Mar. PLoS Pathog. 2024. PMID: 38484003 Free PMC article.
-
β-lactamase-mediated resistance in MDR-Pseudomonas aeruginosa from Qatar.Antimicrob Resist Infect Control. 2020 Nov 1;9(1):170. doi: 10.1186/s13756-020-00838-y. Antimicrob Resist Infect Control. 2020. PMID: 33131487 Free PMC article.
References
-
- Marvig RL, Sommer LM, Jelsbak L, Molin S, Johansen HK. Evolutionary insight from whole-genome sequencing of Pseudomonas aeruginosa from cystic fibrosis patients. Future Microbiol. 2015;10. - PubMed
-
- Feliziani S, Marvig RL, Lujan AM, Moyano AJ, Di Rienzo JA, Krogh Johansen H, et al. Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable Pseudomonas aeruginosa in Long-term Cystic Fibrosis Infections. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004651. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

