Integrated analysis of the local and systemic changes preceding the development of post-partum cytological endometritis
- PMID: 26482908
- PMCID: PMC4617749
- DOI: 10.1186/s12864-015-1967-5
Integrated analysis of the local and systemic changes preceding the development of post-partum cytological endometritis
Erratum in
-
Erratum to: 'Integrated analysis of the local and systemic changes preceding the development of post-partum cytological endometritis'.BMC Genomics. 2015 Dec 10;16:1050. doi: 10.1186/s12864-015-2205-x. BMC Genomics. 2015. PMID: 26653414 Free PMC article. No abstract available.
Abstract
Background: The regulation of endometrial inflammation has important consequences for the resumption of bovine fertility postpartum. All cows experience bacterial influx into the uterus after calving; however a significant proportion fail to clear infection leading to the development of cytological endometritis (CE) and compromised fertility. We hypothesised that early immunological changes could not only act as potential prognostic biomarkers for the subsequent development of disease but also shed light on the pathogenesis of endometritis in the postpartum dairy cow.
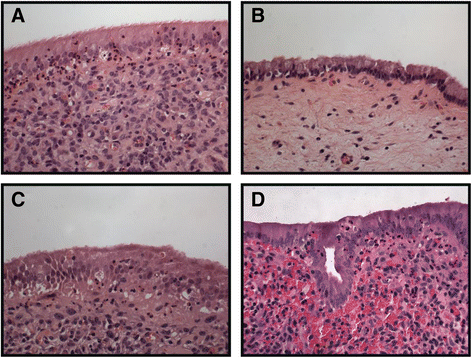
Methods: Endometrial biopsy RNA was extracted from 15 cows at 7 and 21 days postpartum (DPP), using the Qiagen RNeasy(®) Plus Mini kit and quality determined using an Agilent 2100 bioanalyser. Disease status was determined by histpathology based on inflammatory cell infiltrate. RNA-seq of both mRNA and miRNA libraries were performed on an Illumina® HiSeq(™) 2000. Paired reads were aligned to the bovine genome with Bowtie2 and differentially expressed genes were identified using EdgeR. Significantly over-represented Gene Ontology terms were identified using GO-seq, and pathway analysis was performed using KEGG. Quanititative real-time PCR was also performed for validation (ABI 7500 fast). Haematology was assessed using an automated ADVIA 2120 analyser. Serum proteins were evaluated by ELISA and metabolite analysis was performed using a Beckman Coulter AU 400 clinical analyser. Terminal-restriction fragment length polymorphism (T-RFLP) was used to obtain fingerprints of the microbial communities present.
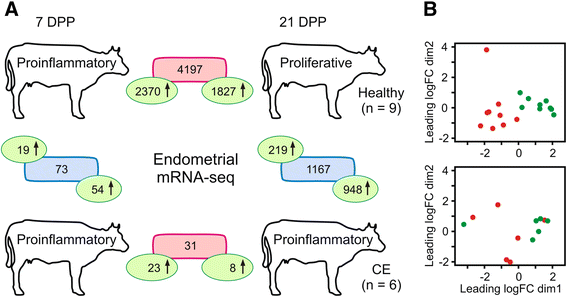
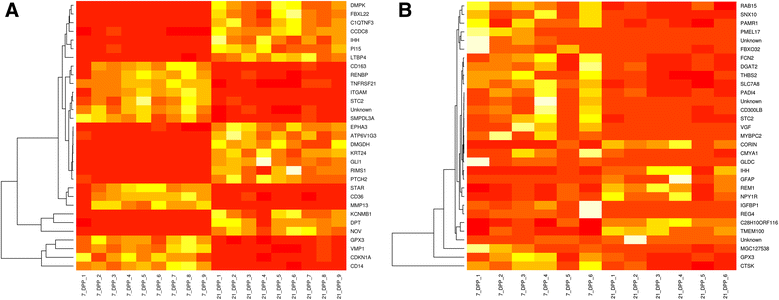
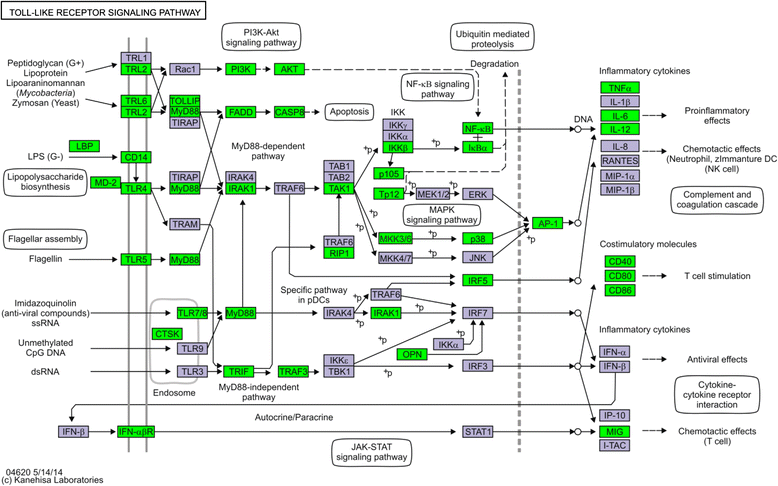
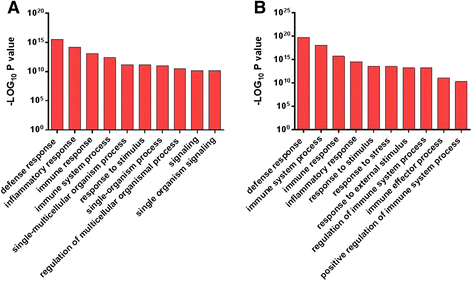
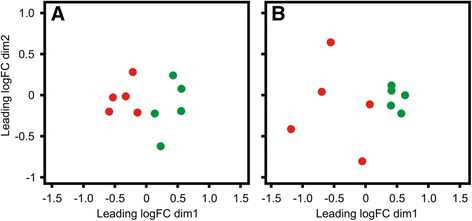
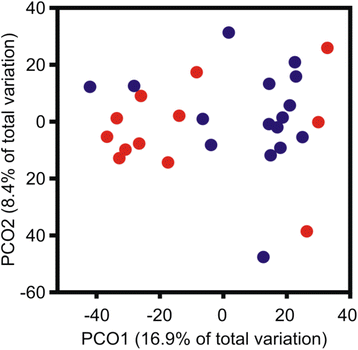
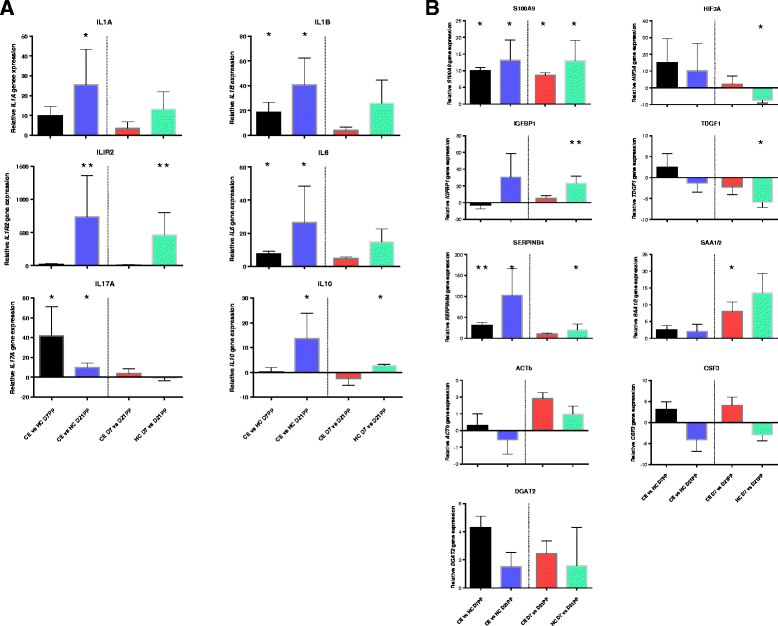
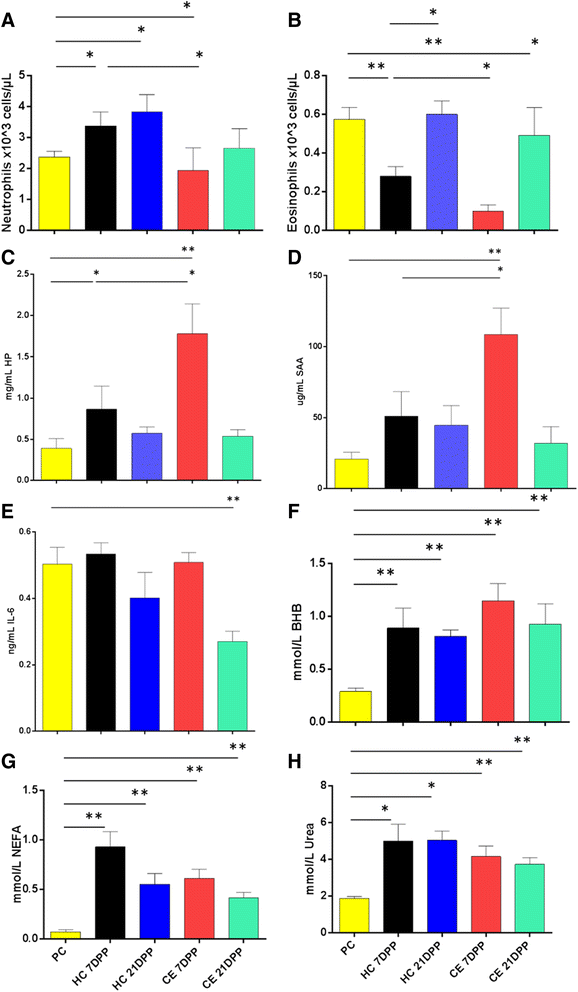
Results: Next-generation sequencing from endometrial biopsies taken at 7 DPP identified significant induction of inflammatory gene expression in all cows. Despite the common inflammatory profile and enrichment of the Toll-like receptor and NFκB pathways, 73 genes and 31 miRNAs were significantly differentially expressed between healthy cows (HC, n = 9) and cows which subsequently developed CE at 7 DPP (n = 6, FDR < 0.1). While significant differential expression of 4197 genes in the transcriptome of healthy cows between 7 and 21 DPP showed the transition from a proinflammatory to tissue profliferation and repair, only 31 genes were differentially expressed in cows with CE (FDR < 0.1), indicating the arrest of such a transition. A link betwene the dysregulated inflammatory response and the composition of the uterine microbial communities was suggested by the presence of significant differences in uterine bacterial tRFLP profiles between HC and CE groups. Furthermore, inflammatory activity was not confined to the uterus; decreased circulating granulocytes and increased Acute Phase Protein (SAA and HP) expression levels were detected in plasma at 7 DPP in cows that developed CE.
Conclusion: Our data suggests that the IL1 and IL17 inflammatory cascade activated early postpartum is resolved thereby restoring homeostasis in healthy cows by 21 DPP, but this transition fails to occur in cows which develop CE. Despite a common early inflammatory profile, elevated and differential expression of specific immune genes may identify cows at risk of prolonged inflammation and the development of CE postpartum.
Figures
References
-
- Kiracofe GH. Uterine involution: its role in regulating postpartum intervals. J Anim Sci. 1980;51(Suppl 2):16–28. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

