Mass spectrometry-based ligand binding assays on adenosine A1 and A2A receptors
- PMID: 26482925
- PMCID: PMC4648803
- DOI: 10.1007/s11302-015-9477-0
Mass spectrometry-based ligand binding assays on adenosine A1 and A2A receptors
Abstract
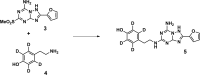
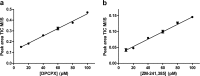
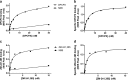
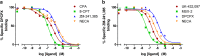
Conventional methods to measure ligand-receptor binding parameters typically require radiolabeled ligands as probes. Despite the robustness of radioligand binding assays, they carry inherent disadvantages in terms of safety precautions, expensive synthesis, special lab requirements, and waste disposal. Mass spectrometry (MS) is a method that can selectively detect ligands without the need of a label. The sensitivity of MS equipment increases progressively, and currently, it is possible to detect low ligand quantities that are usually found in ligand binding assays. We developed a label-free MS ligand binding (MS binding) assay on the adenosine A(1) and A(2A) receptors (A(1)AR and A(2A)AR), which are well-characterized members of the class A G protein-coupled receptor (GPCR) family. Radioligand binding assays for both receptors are well established, and ample data is available to compare and evaluate the performance of an MS binding assay. 1,3-Dipropyl-8-cyclopentyl-xanthine (DPCPX) and 4-(2-((7-amino-2-(furan-2-yl)-[1,2,4]triazolo[1,5-a]-[1,3,5]triazin-5-yl)amino)ethyl)phenol (ZM-241,385) are high-affinity ligands selective for the A(1)AR and A(2A)AR, respectively. To proof the feasibility of MS binding on the A(1)AR and A(2A)AR, we first developed an MS detection method for unlabeled DPCPX and ZM-241,385. To serve as internal standards, both compounds were also deuterium-labeled. Subsequently, we investigated whether the two unlabeled compounds could substitute for their radiolabeled counterparts as marker ligands in binding experiments, including saturation, displacement, dissociation, and competition association assays. Furthermore, we investigated the accuracy of these assays if the use of internal standards was excluded. The results demonstrate the feasibility of the MS binding assay, even in the absence of a deuterium-labeled internal standard, and provide great promise for the further development of label-free assays based on MS for other GPCRs.
Keywords: Adenosine receptor; Deuteration; MS binding; Mass spectrometry; Radioligand binding.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

