RNA Helicase Important for Listeria monocytogenes Hemolytic Activity and Virulence Factor Expression
- PMID: 26483402
- PMCID: PMC4693997
- DOI: 10.1128/IAI.00849-15
RNA Helicase Important for Listeria monocytogenes Hemolytic Activity and Virulence Factor Expression
Abstract
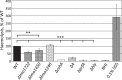
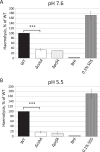
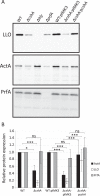
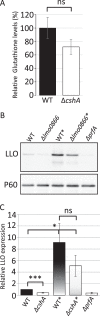
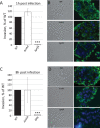
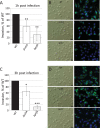
RNA helicases have been shown to be important for the function of RNA molecules at several levels, although their putative involvement in microbial pathogenesis has remained elusive. We have previously shown that Listeria monocytogenes DExD-box RNA helicases are important for bacterial growth, motility, ribosomal maturation, and rRNA processing. We assessed the importance of the RNA helicase Lmo0866 (here named CshA) for expression of virulence traits. We observed a reduction in hemolytic activity in a strain lacking CshA compared to the wild type. This phenomenon was less evident in strains lacking other RNA helicases. The reduced hemolysis was accompanied by lower expression of major listerial virulence factors in the ΔcshA strain, mainly listeriolysin O, but also to some degree the actin polymerizing factor ActA. Reduced expression of these virulence factors in the strain lacking CshA did not, however, correlate with a decreased level of the virulence regulator PrfA. When combining the ΔcshA knockout with a mutation creating a constitutively active PrfA protein (PrfA*), the effect of the ΔcshA knockout on LLO expression was negated. These data suggest a role for the RNA helicase CshA in posttranslational activation of PrfA. Surprisingly, although the expression of several virulence factors was reduced, the ΔcshA strain did not demonstrate any reduced ability to infect nonphagocytic cells compared to the wild-type strain.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
DExD-box RNA-helicases in Listeria monocytogenes are important for growth, ribosomal maturation, rRNA processing and virulence factor expression.RNA Biol. 2014;11(11):1457-66. doi: 10.1080/15476286.2014.996099. RNA Biol. 2014. PMID: 25590644 Free PMC article.
-
Spontaneous Loss of Virulence in Natural Populations of Listeria monocytogenes.Infect Immun. 2017 Oct 18;85(11):e00541-17. doi: 10.1128/IAI.00541-17. Print 2017 Nov. Infect Immun. 2017. PMID: 28827366 Free PMC article.
-
Transcriptional regulation of prfA and PrfA-regulated virulence genes in Listeria monocytogenes.Mol Microbiol. 1994 Mar;11(6):1141-50. doi: 10.1111/j.1365-2958.1994.tb00390.x. Mol Microbiol. 1994. PMID: 8022283
-
Studies on the pathogenicity of Listeria monocytogenes.Infection. 1991;19 Suppl 4:S195-7. doi: 10.1007/BF01644032. Infection. 1991. PMID: 1908834 Review.
-
Flick of a switch: regulatory mechanisms allowing Listeria monocytogenes to transition from a saprophyte to a killer.Microbiology (Reading). 2019 Aug;165(8):819-833. doi: 10.1099/mic.0.000808. Epub 2019 May 20. Microbiology (Reading). 2019. PMID: 31107205 Review.
Cited by
-
Genomic analysis of Indian strains of Salmonella enterica subsp. enterica serovar Typhi indicates novel genetic repertoire for pathogenicity and adaptations.Mol Biol Rep. 2019 Aug;46(4):3967-3989. doi: 10.1007/s11033-019-04843-2. Epub 2019 May 14. Mol Biol Rep. 2019. PMID: 31089918
-
Cold-Shock Domain Family Proteins (Csps) Are Involved in Regulation of Virulence, Cellular Aggregation, and Flagella-Based Motility in Listeria monocytogenes.Front Cell Infect Microbiol. 2017 Oct 26;7:453. doi: 10.3389/fcimb.2017.00453. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29124040 Free PMC article.
-
EⅡB Mutation Reduces the Pathogenicity of Listeria monocytogenes by Negatively Regulating Biofilm Formation Ability, Infective Capacity, and Virulence Gene Expression.Vet Sci. 2024 Jul 2;11(7):301. doi: 10.3390/vetsci11070301. Vet Sci. 2024. PMID: 39057985 Free PMC article.
-
RNA- and protein-mediated control of Listeria monocytogenes virulence gene expression.RNA Biol. 2017 May 4;14(5):460-470. doi: 10.1080/15476286.2016.1189069. Epub 2016 May 24. RNA Biol. 2017. PMID: 27217337 Free PMC article. Review.
-
Nonhemolytic Listeria monocytogenes-Prevalence Rate, Reasons Underlying Atypical Phenotype, and Methods for Accurate Hemolysis Assessment.Microorganisms. 2022 Feb 21;10(2):483. doi: 10.3390/microorganisms10020483. Microorganisms. 2022. PMID: 35208937 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

