Surface Polysaccharide Mutants Reveal that Absence of O Antigen Reduces Biofilm Formation of Actinobacillus pleuropneumoniae
- PMID: 26483403
- PMCID: PMC4694004
- DOI: 10.1128/IAI.00912-15
Surface Polysaccharide Mutants Reveal that Absence of O Antigen Reduces Biofilm Formation of Actinobacillus pleuropneumoniae
Abstract
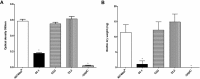
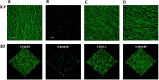
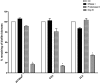
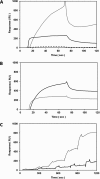
Actinobacillus pleuropneumoniae is a Gram-negative bacterium belonging to the Pasteurellaceae family and the causative agent of porcine pleuropneumonia, a highly contagious lung disease causing important economic losses. Surface polysaccharides, including lipopolysaccharides (LPS) and capsular polysaccharides (CPS), are implicated in the adhesion and virulence of A. pleuropneumoniae, but their role in biofilm formation is still unclear. In this study, we investigated the requirement for these surface polysaccharides in biofilm formation by A. pleuropneumoniae serotype 1. Well-characterized mutants were used: an O-antigen LPS mutant, a truncated core LPS mutant with an intact O antigen, a capsule mutant, and a poly-N-acetylglucosamine (PGA) mutant. We compared the amount of biofilm produced by the parental strain and the isogenic mutants using static and dynamic systems. Compared to the findings for the biofilm of the parental or other strains, the biofilm of the O antigen and the PGA mutants was dramatically reduced, and it had less cell-associated PGA. Real-time PCR analyses revealed a significant reduction in the level of pgaA, cpxR, and cpxA mRNA in the biofilm cells of the O-antigen mutant compared to that in the biofilm cells of the parental strain. Specific binding between PGA and LPS was consistently detected by surface plasmon resonance, but the lack of O antigen did not abolish these interactions. In conclusion, the absence of the O antigen reduces the ability of A. pleuropneumoniae to form a biofilm, and this is associated with the reduced expression and production of PGA.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Identification of genes involved in biosynthesis of Actinobacillus pleuropneumoniae serotype 1 O-antigen and biological properties of rough mutants.J Endotoxin Res. 2002;8(1):27-38. J Endotoxin Res. 2002. PMID: 11981443
-
The CpxA/CpxR Two-Component System Affects Biofilm Formation and Virulence in Actinobacillus pleuropneumoniae.Front Cell Infect Microbiol. 2018 Mar 20;8:72. doi: 10.3389/fcimb.2018.00072. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 29662838 Free PMC article.
-
Isolation and characterization of a capsule-deficient mutant of Actinobacillus pleuropneumoniae serotype 1.Microb Pathog. 2000 May;28(5):279-89. doi: 10.1006/mpat.1999.0347. Microb Pathog. 2000. PMID: 10799278
-
Virulence factors of the swine pathogen Actinobacillus pleuropneumoniae.Microbiologia. 1996 Jun;12(2):171-84. Microbiologia. 1996. PMID: 8767702 Review.
-
Actinobacillus pleuropneumoniae surface polysaccharides: their role in diagnosis and immunogenicity.Anim Health Res Rev. 2000 Dec;1(2):73-93. doi: 10.1017/s1466252300000074. Anim Health Res Rev. 2000. PMID: 11708600 Review.
Cited by
-
Absence of TolC Impairs Biofilm Formation in Actinobacillus pleuropneumoniae by Reducing Initial Attachment.PLoS One. 2016 Sep 28;11(9):e0163364. doi: 10.1371/journal.pone.0163364. eCollection 2016. PLoS One. 2016. PMID: 27681876 Free PMC article.
-
Structural investigation on WlaRG from Campylobacter jejuni: A sugar aminotransferase.Protein Sci. 2017 Mar;26(3):586-599. doi: 10.1002/pro.3109. Epub 2017 Feb 9. Protein Sci. 2017. PMID: 28028852 Free PMC article.
-
Outer Membrane Protein F Is Involved in Biofilm Formation, Virulence and Antibiotic Resistance in Cronobacter sakazakii.Microorganisms. 2021 Nov 11;9(11):2338. doi: 10.3390/microorganisms9112338. Microorganisms. 2021. PMID: 34835462 Free PMC article.
-
Bacterial extracellular matrix as a natural source of biotechnologically multivalent materials.Comput Struct Biotechnol J. 2021 May 5;19:2796-2805. doi: 10.1016/j.csbj.2021.05.008. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34093994 Free PMC article. Review.
-
Why vary what's working? Phase variation and biofilm formation in Francisella tularensis.Front Microbiol. 2022 Dec 6;13:1076694. doi: 10.3389/fmicb.2022.1076694. eCollection 2022. Front Microbiol. 2022. PMID: 36560950 Free PMC article. Review.
References
-
- Gottschalk M. 2012. Actinobacillosis, p 653–669. In Karriker L, Ramirez A, Schwartz K, Stevenson G, Zimmerman J (ed), Diseases of swine, 10th ed John Wiley & Sons, Inc, Hoboken, NJ.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

