Structure of an RNA polymerase II preinitiation complex
- PMID: 26483468
- PMCID: PMC4640751
- DOI: 10.1073/pnas.1518255112
Structure of an RNA polymerase II preinitiation complex
Abstract
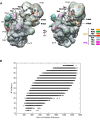
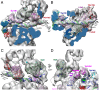
The structure of a 33-protein, 1.5-MDa RNA polymerase II preinitiation complex (PIC) was determined by cryo-EM and image processing at a resolution of 6-11 Å. Atomic structures of over 50% of the mass were fitted into the electron density map in a manner consistent with protein-protein cross-links previously identified by mass spectrometry. The resulting model of the PIC confirmed the main conclusions from previous cryo-EM at lower resolution, including the association of promoter DNA only with general transcription factors and not with the polymerase. Electron density due to DNA was identifiable by the grooves of the double helix and exhibited sharp bends at points downstream of the TATA box, with an important consequence: The DNA at the downstream end coincides with the DNA in a transcribing polymerase. The structure of the PIC is therefore conducive to promoter melting, start-site scanning, and the initiation of transcription.
Keywords: cryo-EM; general transcription factors; transcription; yeast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




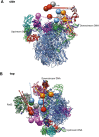




References
-
- Ge H, Roeder RG. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78(3):513–523. - PubMed
-
- Henry NL, Bushnell DA, Kornberg RD. A yeast transcriptional stimulatory protein similar to human PC4. J Biol Chem. 1996;271(36):21842–21847. - PubMed
-
- Brandsen J, et al. C-terminal domain of transcription cofactor PC4 reveals dimeric ssDNA binding site. Nat Struct Biol. 1997;4(11):900–903. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

