Diverse oligomeric states of CEACAM IgV domains
- PMID: 26483485
- PMCID: PMC4640789
- DOI: 10.1073/pnas.1509511112
Diverse oligomeric states of CEACAM IgV domains
Abstract
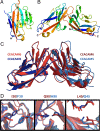
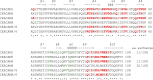
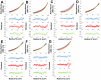
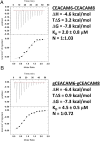
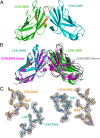
Carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) comprise a large family of cell surface adhesion molecules that bind to themselves and other family members to carry out numerous cellular functions, including proliferation, signaling, differentiation, tumor suppression, and survival. They also play diverse and significant roles in immunity and infection. The formation of CEACAM oligomers is caused predominantly by interactions between their N-terminal IgV domains. Although X-ray crystal structures of CEACAM IgV domain homodimers have been described, how CEACAMs form heterodimers or remain monomers is poorly understood. To address this key aspect of CEACAM function, we determined the crystal structures of IgV domains that form a homodimeric CEACAM6 complex, monomeric CEACAM8, and a heterodimeric CEACAM6-CEACAM8 complex. To confirm and quantify these interactions in solution, we used analytical ultracentrifugation to measure the dimerization constants of CEACAM homodimers and isothermal titration calorimetry to determine the thermodynamic parameters and binding affinities of CEACAM heterodimers. We found the CEACAM6-CEACAM8 heterodimeric state to be substantially favored energetically relative to the CEACAM6 homodimer. Our data provide a molecular basis for the adoption of the diverse oligomeric states known to exist for CEACAMs and suggest ways in which CEACAM6 and CEACAM8 regulate the biological functions of one another, as well as of additional CEACAMs with which they interact, both in cis and in trans.
Keywords: CEACAM; X-ray crystallography; analytical ultracentrifugation; isothermal titration calorimetry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
The role of CEACAMs in neutrophil function.Eur J Clin Invest. 2024 Dec;54 Suppl 2(Suppl 2):e14349. doi: 10.1111/eci.14349. Eur J Clin Invest. 2024. PMID: 39674879 Free PMC article. Review.
-
Carcinoembryonic antigen-related cell adhesion molecule 1 expression and signaling in human, mouse, and rat leukocytes: evidence for replacement of the short cytoplasmic domain isoform by glycosylphosphatidylinositol-linked proteins in human leukocytes.J Immunol. 2002 May 15;168(10):5139-46. doi: 10.4049/jimmunol.168.10.5139. J Immunol. 2002. PMID: 11994468
-
Identification and comparison of residues critical for cell-adhesion activities of two neutrophil CD66 antigens, CEACAM6 and CEACAM8.J Leukoc Biol. 2001 Oct;70(4):543-50. J Leukoc Biol. 2001. PMID: 11590190
-
[Human carcinoembryonic antigen family proteins, structure and function].Postepy Hig Med Dosw (Online). 2012 Jul 20;66:521-33. doi: 10.5604/17322693.1004113. Postepy Hig Med Dosw (Online). 2012. PMID: 22922152 Review. Polish.
-
Binding of Candida albicans to Human CEACAM1 and CEACAM6 Modulates the Inflammatory Response of Intestinal Epithelial Cells.mBio. 2017 Mar 14;8(2):e02142-16. doi: 10.1128/mBio.02142-16. mBio. 2017. PMID: 28292985 Free PMC article.
Cited by
-
Human CEACAM1 N-domain dimerization is independent from glycan modifications.Structure. 2022 May 5;30(5):658-670.e5. doi: 10.1016/j.str.2022.02.003. Epub 2022 Feb 25. Structure. 2022. PMID: 35219398 Free PMC article.
-
Corrigendum: CEACAM1 regulates TIM-3-mediated tolerance and exhaustion.Nature. 2016 Aug 18;536(7616):359. doi: 10.1038/nature17421. Epub 2016 Mar 16. Nature. 2016. PMID: 26982724 Free PMC article. No abstract available.
-
The role of CEACAMs in neutrophil function.Eur J Clin Invest. 2024 Dec;54 Suppl 2(Suppl 2):e14349. doi: 10.1111/eci.14349. Eur J Clin Invest. 2024. PMID: 39674879 Free PMC article. Review.
-
Neisserial Opa Protein-CEACAM Interactions: Competition for Receptors as a Means of Bacterial Invasion and Pathogenesis.Biochemistry. 2016 Aug 9;55(31):4286-94. doi: 10.1021/acs.biochem.6b00124. Epub 2016 Aug 1. Biochemistry. 2016. PMID: 27442026 Free PMC article.
-
Glycosylation Alters Dimerization Properties of a Cell-surface Signaling Protein, Carcinoembryonic Antigen-related Cell Adhesion Molecule 1 (CEACAM1).J Biol Chem. 2016 Sep 16;291(38):20085-95. doi: 10.1074/jbc.M116.740050. Epub 2016 Jul 28. J Biol Chem. 2016. PMID: 27471271 Free PMC article.
References
-
- Zebhauser R, et al. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine Cea family. Genomics. 2005;86(5):566–580. - PubMed
-
- Hammarström S. The carcinoembryonic antigen (CEA) family: Structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9(2):67–81. - PubMed
-
- Singer BB, et al. Carcinoembryonic antigen-related cell adhesion molecule 1 expression and signaling in human, mouse, and rat leukocytes: Evidence for replacement of the short cytoplasmic domain isoform by glycosylphosphatidylinositol-linked proteins in human leukocytes. J Immunol. 2002;168(10):5139–5146. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

