Targeted binding of nucleocapsid protein transforms the folding landscape of HIV-1 TAR RNA
- PMID: 26483503
- PMCID: PMC4640786
- DOI: 10.1073/pnas.1510100112
Targeted binding of nucleocapsid protein transforms the folding landscape of HIV-1 TAR RNA
Abstract
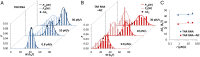
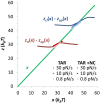
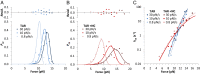
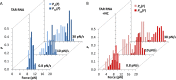
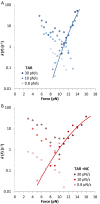
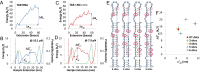
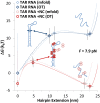
Retroviral nucleocapsid (NC) proteins are nucleic acid chaperones that play a key role in the viral life cycle. During reverse transcription, HIV-1 NC facilitates the rearrangement of nucleic acid secondary structure, allowing the transactivation response (TAR) RNA hairpin to be transiently destabilized and annealed to a cDNA hairpin. It is not clear how NC specifically destabilizes TAR RNA but does not strongly destabilize the resulting annealed RNA-DNA hybrid structure, which must be formed for reverse transcription to continue. By combining single-molecule optical tweezers measurements with a quantitative mfold-based model, we characterize the equilibrium TAR stability and unfolding barrier for TAR RNA. Experiments show that adding NC lowers the transition state barrier height while also dramatically shifting the barrier location. Incorporating TAR destabilization by NC into the mfold-based model reveals that a subset of preferential protein binding sites is responsible for the observed changes in the unfolding landscape, including the unusual shift in the transition state. We measure the destabilization induced at these NC binding sites and find that NC preferentially targets TAR RNA by binding to specific sequence contexts that are not present on the final annealed RNA-DNA hybrid structure. Thus, specific binding alters the entire RNA unfolding landscape, resulting in the dramatic destabilization of this specific structure that is required for reverse transcription.
Keywords: RNA binding; RNA stretching; force spectroscopy; single molecule.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Significant Differences in RNA Structure Destabilization by HIV-1 GagDp6 and NCp7 Proteins.Viruses. 2020 Apr 25;12(5):484. doi: 10.3390/v12050484. Viruses. 2020. PMID: 32344834 Free PMC article.
-
Specific Nucleic Acid Chaperone Activity of HIV-1 Nucleocapsid Protein Deduced from Hairpin Unfolding.Methods Mol Biol. 2020;2106:59-88. doi: 10.1007/978-1-0716-0231-7_4. Methods Mol Biol. 2020. PMID: 31889251
-
Nucleic acid conformational changes essential for HIV-1 nucleocapsid protein-mediated inhibition of self-priming in minus-strand transfer.J Mol Biol. 2003 Jan 3;325(1):1-10. doi: 10.1016/s0022-2836(02)01177-4. J Mol Biol. 2003. PMID: 12473448
-
Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism.Prog Nucleic Acid Res Mol Biol. 2005;80:217-86. doi: 10.1016/S0079-6603(05)80006-6. Prog Nucleic Acid Res Mol Biol. 2005. PMID: 16164976 Review. No abstract available.
-
Nucleocapsid protein function in early infection processes.Virus Res. 2008 Jun;134(1-2):39-63. doi: 10.1016/j.virusres.2007.12.006. Epub 2008 Feb 14. Virus Res. 2008. PMID: 18279991 Free PMC article. Review.
Cited by
-
Force-induced melting and S-DNA pathways for DNA overstretching exhibit distinct kinetics.Nucleic Acids Res. 2025 Jan 7;53(1):gkae1183. doi: 10.1093/nar/gkae1183. Nucleic Acids Res. 2025. PMID: 39657777 Free PMC article.
-
Nucleocapsid Protein Precursors NCp9 and NCp15 Suppress ATP-Mediated Rescue of AZT-Terminated Primers by HIV-1 Reverse Transcriptase.Antimicrob Agents Chemother. 2020 Sep 21;64(10):e00958-20. doi: 10.1128/AAC.00958-20. Print 2020 Sep 21. Antimicrob Agents Chemother. 2020. PMID: 32747359 Free PMC article.
-
Human FACT subunits coordinate to catalyze both disassembly and reassembly of nucleosomes.Cell Rep. 2022 Dec 27;41(13):111858. doi: 10.1016/j.celrep.2022.111858. Cell Rep. 2022. PMID: 36577379 Free PMC article.
-
Retroviral nucleocapsid proteins and DNA strand transfers.Biochim Open. 2018 Jul 20;7:10-25. doi: 10.1016/j.biopen.2018.07.001. eCollection 2018 Dec. Biochim Open. 2018. PMID: 30109196 Free PMC article. Review.
-
The Life-Cycle of the HIV-1 Gag-RNA Complex.Viruses. 2016 Sep 10;8(9):248. doi: 10.3390/v8090248. Viruses. 2016. PMID: 27626439 Free PMC article. Review.
References
-
- Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog Nucleic Acid Res Mol Biol. 2005;80:217–286. - PubMed
-
- Vo MN, Barany G, Rouzina I, Musier-Forsyth K. Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J Mol Biol. 2006;363(1):244–261. - PubMed
-
- Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC. Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein’s nucleic acid chaperone function. J Mol Biol. 2006;363(5):867–877. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

