Salt Bridge Swapping in the EXXERFXYY Motif of Proton-coupled Oligopeptide Transporters
- PMID: 26483552
- PMCID: PMC4705975
- DOI: 10.1074/jbc.M115.675603
Salt Bridge Swapping in the EXXERFXYY Motif of Proton-coupled Oligopeptide Transporters
Abstract
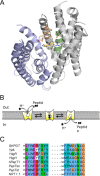
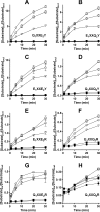
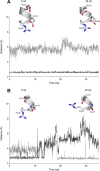
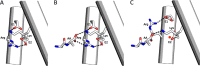
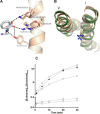
Proton-coupled oligopeptide transporters (POTs) couple the inward transport of di- or tripeptides with an inwardly directed transport of protons. Evidence from several studies of different POTs has pointed toward involvement of a highly conserved sequence motif, E1XXE2RFXYY (from here on referred to as E1XXE2R), located on Helix I, in interactions with the proton. In this study, we investigated the intracellular substrate accumulation by motif variants with all possible combinations of glutamate residues changed to glutamine and arginine changed to a tyrosine, the latter being a natural variant found in the Escherichia coli POT YjdL. We found that YjdL motif variants with E1XXE2R, E1XXE2Y, E1XXQ2Y, or Q1XXE2Y were able to accumulate peptide, whereas those with E1XXQ2R, Q1XXE2R, or Q1XXQ2Y were unable to accumulate peptide, and Q1XXQ2R abolished uptake. These results suggest a mechanism that involves swapping of an intramotif salt bridge, i.e. R-E2 to R-E1, which is consistent with previous structural studies. Molecular dynamics simulations of the motif variants E1XXE2R and E1XXQ2R support this mechanism. The simulations showed that upon changing conformation arginine pushes Helix V, through interactions with the highly conserved FYING motif, further away from the central cavity in what could be a stabilization of an inward facing conformation. As E2 has been suggested to be the primary site for protonation, these novel findings show how protonation may drive conformational changes through interactions of two highly conserved motifs.
Keywords: membrane transport; molecular dynamics; peptide transport; protein structure; site-directed mutagenesis.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

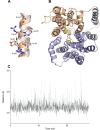
References
-
- Fei Y. J., Kanai Y., Nussberger S., Ganapathy V., Leibach F. H., Romero M. F., Singh S. K., Boron W. F., and Hediger M. A. (1994) Expression cloning of a mammalian proton-coupled oligopeptide transporter. Nature 368, 563–566 - PubMed
-
- Liang R., Fei Y. J., Prasad P. D., Ramamoorthy S., Han H., Yang-Feng T. L., Hediger M. A., Ganapathy V., and Leibach F. H. (1995) Human intestinal H+/peptide cotransporter. Cloning, functional expression, and chromosomal localization. J. Biol. Chem. 270, 6456–6463 - PubMed
-
- Weitz D., Harder D., Casagrande F., Fotiadis D., Obrdlik P., Kelety B., and Daniel H. (2007) Functional and structural characterization of a prokaryotic peptide transporter with features similar to mammalian PEPT1. J. Biol. Chem. 282, 2832–2839 - PubMed
-
- Theis S., Döring F., and Daniel H. (2001) Expression of the myc/His-tagged human peptide transporter hPEPT1 in yeast for protein purification and functional analysis. Protein Expr. Purif. 22, 436–442 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

