Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila
- PMID: 26483560
- PMCID: PMC4621839
- DOI: 10.1083/jcb.201502032
Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila
Abstract
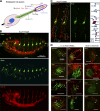
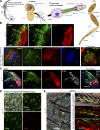
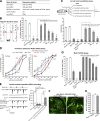
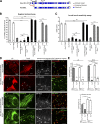
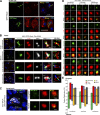
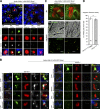
Cilia are essential for cell signaling and sensory perception. In many cell types, a cytoskeletal structure called the ciliary rootlet links the cilium to the cell body. Previous studies indicated that rootlets support the long-term stability of some cilia. Here we report that Drosophila melanogaster Rootletin (Root), the sole orthologue of the mammalian paralogs Rootletin and C-Nap1, assembles into rootlets of diverse lengths among sensory neuron subtypes. Root mutant neurons lack rootlets and have dramatically impaired sensory function, resulting in behavior defects associated with mechanosensation and chemosensation. Root is required for cohesion of basal bodies, but the cilium structure appears normal in Root mutant neurons. We show, however, that normal rootlet assembly requires centrioles. The N terminus of Root contains a conserved domain and is essential for Root function in vivo. Ectopically expressed Root resides at the base of mother centrioles in spermatocytes and localizes asymmetrically to mother centrosomes in neuroblasts, both requiring Bld10, a basal body protein with varied functions.
© 2015 Chen et al.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

