Facilitating the Validation of Novel Protein Biomarkers for Dementia: An Optimal Workflow for the Development of Sandwich Immunoassays
- PMID: 26483753
- PMCID: PMC4586418
- DOI: 10.3389/fneur.2015.00202
Facilitating the Validation of Novel Protein Biomarkers for Dementia: An Optimal Workflow for the Development of Sandwich Immunoassays
Abstract
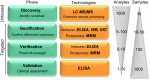
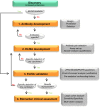
Different neurodegenerative disorders, such as Alzheimer's disease (AD) and frontotemporal dementia (FTD), lead to dementia syndromes. Dementia will pose a huge impact on society and thus it is essential to develop novel tools that are able to detect the earliest, most sensitive, discriminative, and dynamic biomarkers for each of the disorders. To date, the most common assays used in large-scale protein biomarker analysis are enzyme-linked immunosorbent assays (ELISA), such as the sandwich immunoassays, which are sensitive, practical, and easily implemented. However, due to the novelty of many candidate biomarkers identified during proteomics screening, such assays or the antibodies that specifically recognize the desired marker are often not available. The development and optimization of a new ELISA should be carried out with considerable caution since a poor planning can be costly, ineffective, time consuming, and it may lead to a misinterpretation of the findings. Previous guidelines described either the overall biomarker development in more general terms (i.e., the process from biomarker discovery to validation) or the specific steps of performing an ELISA procedure. However, a workflow describing and guiding the main issues in the development of a novel ELISA is missing. Here, we describe a specific and detailed workflow to develop and validate new ELISA for a successful and reliable validation of novel dementia biomarkers. The proposed workflow highlights the main issues in the development of an ELISA and covers several critical aspects, including production, screening, and selection of specific antibodies until optimal fine-tuning of the assay. Although these recommendations are designed to analyze novel biomarkers for dementia in cerebrospinal fluid, they are generally applicable for the development of immunoassays for biomarkers in other human body fluids or tissues. This workflow is designed to maximize the quality of the developed ELISA using a time- and cost-efficient strategy. This will facilitate the validation of the dementia biomarker candidates ultimately allowing accurate diagnostic conclusions.
Keywords: AD; CSF; ELISA; FTD; dementia; guidelines; novel biomarkers; workflow.
Figures


References
-
- Alzheimer’s Disease International. World Alzheimer Report 2010. The Global Economic Impact of Dementia. London: Alzheimer’s disease International; (2010).
-
- Jack CR, Albert MS, Knopman DS, McKhann GM, Sperling RA, Carrillo MC, et al. Introduction to the recommendations from the National Institute on aging-Alzheimer’s association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement (2011) 7:257–62.10.1016/j.jalz.2011.03.004 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

