Evolutionary history influences the salinity preference of bacterial taxa in wetland soils
- PMID: 26483764
- PMCID: PMC4591843
- DOI: 10.3389/fmicb.2015.01013
Evolutionary history influences the salinity preference of bacterial taxa in wetland soils
Abstract
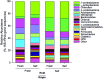
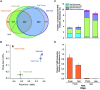
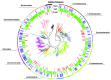
Salinity is a major driver of bacterial community composition across the globe. Despite growing recognition that different bacterial species are present or active at different salinities, the mechanisms by which salinity structures community composition remain unclear. We tested the hypothesis that these patterns reflect ecological coherence in the salinity preferences of phylogenetic groups using a reciprocal transplant experiment of fresh- and saltwater wetland soils. The salinity of both the origin and host environments affected community composition (16S rRNA gene sequences) and activity (CO2 and CH4 production, and extracellular enzyme activity). These changes in community composition and activity rates were strongly correlated, which suggests the effect of environment on function could be mediated, at least in part, by microbial community composition. Based on their distribution across treatments, each phylotype was categorized as having a salinity preference (freshwater, saltwater, or none) and phylogenetic analyses revealed a significant influence of evolutionary history on these groupings. This finding was corroborated by examining the salinity preferences of high-level taxonomic groups. For instance, we found that the majority of α- and γ-proteobacteria in these wetland soils preferred saltwater, while many β-proteobacteria prefer freshwater. Overall, our results indicate the effect of salinity on bacterial community composition results from phylogenetically-clustered salinity preferences.
Keywords: biogeochemistry; community composition; marsh; methanogen; phylogeny; saltwater intrusion.
Figures
Similar articles
-
Novel microbial community composition and carbon biogeochemistry emerge over time following saltwater intrusion in wetlands.Glob Chang Biol. 2019 Feb;25(2):549-561. doi: 10.1111/gcb.14486. Epub 2018 Dec 9. Glob Chang Biol. 2019. PMID: 30537235
-
Salinity affects microbial activity and soil organic matter content in tidal wetlands.Glob Chang Biol. 2014 Apr;20(4):1351-62. doi: 10.1111/gcb.12431. Epub 2014 Jan 13. Glob Chang Biol. 2014. PMID: 24307658
-
Phosphorus alleviation of salinity stress: effects of saltwater intrusion on an Everglades freshwater peat marsh.Ecology. 2019 May;100(5):e02672. doi: 10.1002/ecy.2672. Epub 2019 Apr 3. Ecology. 2019. PMID: 30942486
-
Salinity tolerances and use of saline environments by freshwater turtles: implications of sea level rise.Biol Rev Camb Philos Soc. 2018 Aug;93(3):1634-1648. doi: 10.1111/brv.12410. Epub 2018 Mar 25. Biol Rev Camb Philos Soc. 2018. PMID: 29575680 Review.
-
A Review of Salinity's Impact on Relevant Aquatic Drug Approval Technical Sections.J Aquat Anim Health. 2022 Sep;34(3):149-155. doi: 10.1002/aah.10168. J Aquat Anim Health. 2022. PMID: 36165570 Review.
Cited by
-
Salt marsh sediment bacterial communities maintain original population structure after transplantation across a latitudinal gradient.PeerJ. 2018 May 1;6:e4735. doi: 10.7717/peerj.4735. eCollection 2018. PeerJ. 2018. PMID: 29736349 Free PMC article.
-
Trace Metal Availability Affects Greenhouse Gas Emissions and Microbial Functional Group Abundance in Freshwater Wetland Sediments.Front Microbiol. 2020 Sep 30;11:560861. doi: 10.3389/fmicb.2020.560861. eCollection 2020. Front Microbiol. 2020. PMID: 33117308 Free PMC article.
-
Bacterial community response to novel and repeated disturbances.Environ Microbiol Rep. 2024 Oct;16(5):e70022. doi: 10.1111/1758-2229.70022. Environ Microbiol Rep. 2024. PMID: 39387551 Free PMC article.
-
Changes in sediment greenhouse gases production dynamics in an estuarine wetland following invasion by Spartina alterniflora.Front Microbiol. 2024 Jul 12;15:1420924. doi: 10.3389/fmicb.2024.1420924. eCollection 2024. Front Microbiol. 2024. PMID: 39070262 Free PMC article.
-
The Microbiome of Neotropical Water Striders and Its Potential Role in Codiversification.Insects. 2020 Aug 31;11(9):578. doi: 10.3390/insects11090578. Insects. 2020. PMID: 32878094 Free PMC article.
References
-
- Balser T. C., Firestone M. K. (2005). Linking microbial community composition and soil processes in a California annual grassland and mixed-conifer forest. Biogeochemistry 73 395–415. 10.1007/s10533-004-0372-y - DOI
-
- Bartram A. K., Lynch M. D. J., Stearns J. C., Moreno-Hagelsieb G., Neufeld J. D. (2011). Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl. Environ. Microbiol. 77 3846–3852. 10.1128/AEM.02772-10 - DOI - PMC - PubMed
-
- Berner R. A. (1970). Sedimentary pyrite formation. Am. J. Sci. 268 1–23. 10.2475/ajs.268.1.1 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

