Lassa virus isolates from Mali and the Ivory Coast represent an emerging fifth lineage
- PMID: 26483768
- PMCID: PMC4589679
- DOI: 10.3389/fmicb.2015.01037
Lassa virus isolates from Mali and the Ivory Coast represent an emerging fifth lineage
Abstract
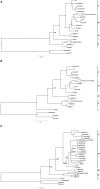
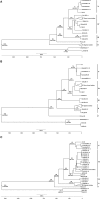
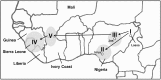
Previous imported cases of Lassa fever (LF) into the United Kingdom from the Ivory Coast and Mali, as well as the detection of Lassa virus (LASV) among the Mastomys natalensis population within Mali has led to the suggestion that the endemic area for LF is expanding. Initial phylogenetic analyses arrange isolates from Mali and the Ivory Coast separately from the classical lineage IV isolates taken from Sierra Leone, Guinea, and Liberia. The availability of full genome sequences continues to increase, allowing for a more complete phylogenetic comparison of the isolates from Mali and the Ivory Coast to the other existing isolates. In this study, we utilized a Bayesian approach to infer the demographic histories of each LASV isolate for which the full sequence was available. Our results indicate that the isolates from Mali and the Ivory Coast group separately from the isolates of lineage IV, comprising a distinct fifth lineage. The split between lineages IV and V is estimated to have occurred around 200-300 years ago, which coincides with the colonial period of West Africa.
Keywords: Lassa fever; Lassa virus; genetic diversity; lineage; phylogenetics.
Figures
References
-
- Atkin S., Anaraki S., Gothard P., Walsh A., Brown D., Gopal R., et al. . (2009). The first case of Lassa fever imported from Mali to the United Kingdom, February 2009. Euro Surveill. 14:19145. Available online at: http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19145 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

