Choosing a suitable method for the identification of replication origins in microbial genomes
- PMID: 26483774
- PMCID: PMC4588119
- DOI: 10.3389/fmicb.2015.01049
Choosing a suitable method for the identification of replication origins in microbial genomes
Abstract
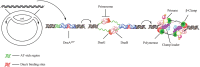
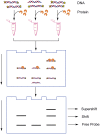
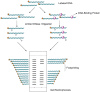
As the replication of genomic DNA is arguably the most important task performed by a cell and given that it is controlled at the initiation stage, the events that occur at the replication origin play a central role in the cell cycle. Making sense of DNA replication origins is important for improving our capacity to study cellular processes and functions in the regulation of gene expression, genome integrity in much finer detail. Thus, clearly comprehending the positions and sequences of replication origins which are fundamental to chromosome organization and duplication is the first priority of all. In view of such important roles of replication origins, tremendous work has been aimed at identifying and testing the specificity of replication origins. A number of computational tools based on various skew types have been developed to predict replication origins. Using various in silico approaches such as Ori-Finder, and databases such as DoriC, researchers have predicted the locations of replication origins sites for thousands of bacterial chromosomes and archaeal genomes. Based on the predicted results, we should choose an effective method for identifying and confirming the interactions at origins of replication. Here we describe the main existing experimental methods that aimed to determine the replication origin regions and list some of the many the practical applications of these methods.
Keywords: ChIP; ChIP-seq; Dnase I footprinting; EMSA; ITC; RIP mapping; SPR; replication origin.
Figures
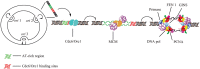
Similar articles
-
Recent development of Ori-Finder system and DoriC database for microbial replication origins.Brief Bioinform. 2019 Jul 19;20(4):1114-1124. doi: 10.1093/bib/bbx174. Brief Bioinform. 2019. PMID: 29329409 Review.
-
DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes.Nucleic Acids Res. 2013 Jan;41(Database issue):D90-3. doi: 10.1093/nar/gks990. Epub 2012 Oct 23. Nucleic Acids Res. 2013. PMID: 23093601 Free PMC article.
-
DoriC 10.0: an updated database of replication origins in prokaryotic genomes including chromosomes and plasmids.Nucleic Acids Res. 2019 Jan 8;47(D1):D74-D77. doi: 10.1093/nar/gky1014. Nucleic Acids Res. 2019. PMID: 30364951 Free PMC article.
-
Ori-Finder 2, an integrated tool to predict replication origins in the archaeal genomes.Front Microbiol. 2014 Sep 15;5:482. doi: 10.3389/fmicb.2014.00482. eCollection 2014. Front Microbiol. 2014. PMID: 25309521 Free PMC article.
-
Identification of replication origins in prokaryotic genomes.Brief Bioinform. 2008 Sep;9(5):376-91. doi: 10.1093/bib/bbn031. Epub 2008 Jul 26. Brief Bioinform. 2008. PMID: 18660512 Review.
Cited by
-
Biophysical and biochemical insights in the design of immunoassays.Biochim Biophys Acta Gen Subj. 2023 Jan;1867(1):130266. doi: 10.1016/j.bbagen.2022.130266. Epub 2022 Oct 26. Biochim Biophys Acta Gen Subj. 2023. PMID: 36309294 Free PMC article. Review.
-
Ori-Finder 2022: A Comprehensive Web Server for Prediction and Analysis of Bacterial Replication Origins.Genomics Proteomics Bioinformatics. 2022 Dec;20(6):1207-1213. doi: 10.1016/j.gpb.2022.10.002. Epub 2022 Oct 17. Genomics Proteomics Bioinformatics. 2022. PMID: 36257484 Free PMC article.
-
The Epigenetics of Anxiety Pathophysiology: A DNA Methylation and Histone Modification Focused Review.eNeuro. 2023 Apr 24;10(4):ENEURO.0109-21.2021. doi: 10.1523/ENEURO.0109-21.2021. Print 2023 Apr. eNeuro. 2023. PMID: 35998298 Free PMC article. Review.
-
gammaBOriS: Identification and Taxonomic Classification of Origins of Replication in Gammaproteobacteria using Motif-based Machine Learning.Sci Rep. 2020 Apr 21;10(1):6727. doi: 10.1038/s41598-020-63424-7. Sci Rep. 2020. PMID: 32317695 Free PMC article.
-
Computational prediction of species-specific yeast DNA replication origin via iterative feature representation.Brief Bioinform. 2021 Jul 20;22(4):bbaa304. doi: 10.1093/bib/bbaa304. Brief Bioinform. 2021. PMID: 33232970 Free PMC article.
References
-
- Aves S. J. (2009). “DNA replication initiation,” in DNA Replication eds Vengrova S., Dalgaard J. Z. (New York, NY: Humana Press; ) 1–16. 10.1093/jexbot/52.355.193 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

