Probing the regulatory effects of specific mutations in three major binding domains of the pleiotropic regulator CcpA of Bacillus subtilis
- PMID: 26483775
- PMCID: PMC4591507
- DOI: 10.3389/fmicb.2015.01051
Probing the regulatory effects of specific mutations in three major binding domains of the pleiotropic regulator CcpA of Bacillus subtilis
Abstract
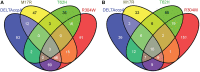
Carbon catabolite control is required for efficient use of available carbon sources to ensure rapid growth of bacteria. CcpA is a global regulator of carbon metabolism in Gram-positive bacteria like Bacillus subtilis. In this study the genome-wide gene regulation of a CcpA knockout and three specific CcpA mutants were studied by transcriptome analysis, to further elucidate the function of specific binding sites in CcpA. The following three amino acids were mutated to characterize their function: M17(R) which is involved in DNA binding, T62(H) which is important for the allosteric switch in CcpA upon HPr-Ser46-P binding, and R304(W) which is important for binding of the coeffectors HPr-Ser46-P and fructose-1,6-bisphosphate. The results confirm that CcpA was also involved in gene regulation in the absence of glucose. CcpA-M17R showed a small relief of Carbon Catabolite Control; the CcpA-M17R mutant regulates fewer genes than the CcpA-wt and the palindromicity of the cre site is less important for CcpA-M17R. CcpA-T62H was a stronger repressor than CcpA-wt and also acted as a strong repressor in the absence of glucose. CcpA-R304W was shown here to be less dependent on HPr-Ser46-P for its carbon catabolite control activities. The results presented here provide detailed information on alterations in gene regulation for each CcpA-mutant.
Keywords: Bacillus subtilis; CcpA; CcpA mutants; DNA microarray; transcriptomics.
Figures


Similar articles
-
Residues His-15 and Arg-17 of HPr participate differently in catabolite signal processing via CcpA.J Biol Chem. 2007 Jan 12;282(2):1175-82. doi: 10.1074/jbc.M605854200. Epub 2006 Nov 3. J Biol Chem. 2007. PMID: 17085448
-
Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.Nucleic Acids Res. 2011 Apr;39(7):2931-42. doi: 10.1093/nar/gkq1177. Epub 2010 Nov 23. Nucleic Acids Res. 2011. PMID: 21106498 Free PMC article.
-
Phosphoprotein Crh-Ser46-P displays altered binding to CcpA to effect carbon catabolite regulation.J Biol Chem. 2006 Mar 10;281(10):6793-800. doi: 10.1074/jbc.M509977200. Epub 2005 Nov 29. J Biol Chem. 2006. PMID: 16316990
-
Catabolite repression in Bacillus subtilis: a global regulatory mechanism for the gram-positive bacteria?Mol Microbiol. 1995 Feb;15(3):395-401. doi: 10.1111/j.1365-2958.1995.tb02252.x. Mol Microbiol. 1995. PMID: 7540244 Review.
-
CcpA-independent carbon catabolite repression in Bacillus subtilis.J Mol Microbiol Biotechnol. 2002 May;4(3):315-21. J Mol Microbiol Biotechnol. 2002. PMID: 11931564 Review.
Cited by
-
Understanding and application of Bacillus nitrogen regulation: A synthetic biology perspective.J Adv Res. 2023 Jul;49:1-14. doi: 10.1016/j.jare.2022.09.003. Epub 2022 Sep 12. J Adv Res. 2023. PMID: 36103961 Free PMC article. Review.
-
A new CcpA binding site plays a bidirectional role in carbon catabolism in Bacillus licheniformis.iScience. 2021 Apr 7;24(5):102400. doi: 10.1016/j.isci.2021.102400. eCollection 2021 May 21. iScience. 2021. PMID: 33997685 Free PMC article.
-
Microbial cell factories for bio-based biodegradable plastics production.iScience. 2022 Oct 31;25(11):105462. doi: 10.1016/j.isci.2022.105462. eCollection 2022 Nov 18. iScience. 2022. PMID: 36405773 Free PMC article. Review.
References
-
- Detert Oude Weme R. G., Kovacs A. T., de Jong S. J., Veening J. W., Siebring J., Kuipers O. P. (2015). Single cell FRET analysis for the identification of optimal FRET-pairs in Bacillus subtilis using a prototype MEM-FLIM system. PLoS ONE 10:e0123239 10.1371/journal.pone.0123239 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

