Recovering full-length viral genomes from metagenomes
- PMID: 26483782
- PMCID: PMC4589665
- DOI: 10.3389/fmicb.2015.01069
Recovering full-length viral genomes from metagenomes
Abstract
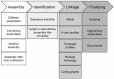
Infectious disease metagenomics is driven by the question: "what is causing the disease?" in contrast to classical metagenome studies which are guided by "what is out there?" In case of a novel virus, a first step to eventually establishing etiology can be to recover a full-length viral genome from a metagenomic sample. However, retrieval of a full-length genome of a divergent virus is technically challenging and can be time-consuming and costly. Here we discuss different assembly and fragment linkage strategies such as iterative assembly, motif searches, k-mer frequency profiling, coverage profile binning, and other strategies used to recover genomes of potential viral pathogens in a timely and cost-effective manner.
Keywords: assembly; coverage analysis; k-mer analysis; metagenomics; motif discovery; virus discovery; viruses; zoonotic pathogens.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

