A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.)
- PMID: 26483801
- PMCID: PMC4588698
- DOI: 10.3389/fpls.2015.00770
A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.)
Abstract
The Auxin/indole-3-acetic acid (Aux/IAA) gene family plays key roles in the primary auxin-response process and controls a number of important traits in plants. However, the characteristics of the Aux/IAA gene family in hexaploid bread wheat (Triticum aestivum L.) have long been unknown. In this study, a comprehensive identification of the Aux/IAA gene family was performed using the latest draft genome sequence of the bread wheat "Chinese Spring." Thirty-four Aux/IAA genes were identified, 30 of which have duplicated genes on the A, B or D sub-genome, with a total of 84 Aux/IAA sequences. These predicted Aux/IAA genes were non-randomly distributed in all the wheat chromosomes except for chromosome 2D. The information of wheat Aux/IAA proteins is also described. Based on an analysis of phylogeny, expression and adaptive evolution, we prove that the Aux/IAA family in wheat has been replicated twice in the two allopolyploidization events of bread wheat, when the tandem duplication also occurred. The duplicated genes have undergone an evolutionary process of purifying selection, resulting in the high conservation of copy genes among sub-genomes and functional redundancy among several members of the TaIAA family. However, functional divergence probably existed in most TaIAA members due to the diversity of the functional domain and expression pattern. Our research provides useful information for further research into the function of Aux/IAA genes in wheat.
Keywords: Aux/IAA family; bread wheat genome; chromosome location; expansion pattern; function prediction.
Figures


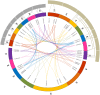
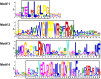


References
-
- Akhunov E. D., Sehgal S., Liang H., Wang S., Akhunova A. R., Kaur G., et al. . (2013). Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 161, 252–265. 10.1104/pp.112.205161 - DOI - PMC - PubMed
-
- Birsen C., Ozan K., Ahmet C. O. (2013). Genome-wide analysis of Aux/IAA genes in Vitis vinifera: cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses. Acta. Physiol. Plant. 35, 365–377. 10.1007/s11738-012-1079-7 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

