Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress
- PMID: 26483820
- PMCID: PMC4589653
- DOI: 10.3389/fpls.2015.00806
Genome-wide analysis of the CaHsp20 gene family in pepper: comprehensive sequence and expression profile analysis under heat stress
Abstract
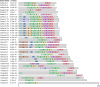
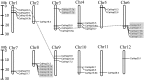
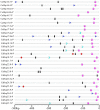
The Hsp20 genes are present in all plant species and play important roles in alleviating heat stress and enhancing plant thermotolerance by preventing the irreversible aggregation of denaturing proteins. However, very little is known about the CaHsp20 gene family in pepper (Capsicum annuum L.), an important vegetable crop with character of temperate but thermosensitive. In this study, a total of 35 putative pepper Hsp20 genes (CaHsp20s) were identified and renamed on the basis of their molecular weight, and then their gene structure, genome location, gene duplication, phylogenetic relationship, and interaction network were also analyzed. The expression patterns of CaHsp20 genes in four different tissues (root, stem, leaf, and flower) from the thermotolerant line R9 under heat stress condition were measured using semi-quantitative RT-PCR. The transcripts of most CaHsp20 genes maintained a low level in all of the four tissues under normal temperature condition, but were highly induced by heat stress, while the expression of CaHsp16.6b, 16.7, and 23.8 were only detected in specific tissues and were not so sensitive to heat stress like other CaHsp20 genes. In addition, compared to those in thermotolerant line R9, the expression peak of most CaHsp20 genes in thermosensitive line B6 under heat stress was hysteretic, and several CaHsp20 genes (CaHsp16.4, 18.2a, 18.7, 21.2, 22.0, 25.8, and 25.9) showed higher expression levels in both line B6 and R9. These data suggest that the CaHsp20 genes may be involved in heat stress and defense responses in pepper, which provides the basis for further functional analyses of CaHsp20s in the formation of pepper acquired thermotoleance.
Keywords: CaHsp20 genes; Capsicum annuum L.; expression profile; gene family; heat stress.
Figures





Similar articles
-
CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance.Protoplasma. 2019 Jan;256(1):39-51. doi: 10.1007/s00709-018-1280-7. Epub 2018 Jun 26. Protoplasma. 2019. PMID: 29946904
-
Genome-wide analysis, expression profile of heat shock factor gene family (CaHsfs) and characterisation of CaHsfA2 in pepper (Capsicum annuum L.).BMC Plant Biol. 2015 Jun 19;15:151. doi: 10.1186/s12870-015-0512-7. BMC Plant Biol. 2015. PMID: 26088319 Free PMC article.
-
Characterization of CaHsp70-1, a pepper heat-shock protein gene in response to heat stress and some regulation exogenous substances in Capsicum annuum L.Int J Mol Sci. 2014 Oct 29;15(11):19741-59. doi: 10.3390/ijms151119741. Int J Mol Sci. 2014. PMID: 25356507 Free PMC article.
-
Genome-Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis-Elements in the Promoter, and Interaction Network.Int J Mol Sci. 2018 Mar 29;19(4):1028. doi: 10.3390/ijms19041028. Int J Mol Sci. 2018. PMID: 29596349 Free PMC article.
-
Final report on the safety assessment of capsicum annuum extract, capsicum annuum fruit extract, capsicum annuum resin, capsicum annuum fruit powder, capsicum frutescens fruit, capsicum frutescens fruit extract, capsicum frutescens resin, and capsaicin.Int J Toxicol. 2007;26 Suppl 1:3-106. doi: 10.1080/10915810601163939. Int J Toxicol. 2007. PMID: 17365137 Review.
Cited by
-
Genome-wide identification, characterization and expression of HSP 20 gene family in dove.Front Genet. 2022 Oct 4;13:1011676. doi: 10.3389/fgene.2022.1011676. eCollection 2022. Front Genet. 2022. PMID: 36267407 Free PMC article.
-
Identification of the DcHsp20 gene family in carnation (Dianthus caryophyllus) and functional characterization of DcHsp17.8 in heat tolerance.Planta. 2022 May 27;256(1):2. doi: 10.1007/s00425-022-03915-1. Planta. 2022. PMID: 35624182
-
CaHSP16.4, a small heat shock protein gene in pepper, is involved in heat and drought tolerance.Protoplasma. 2019 Jan;256(1):39-51. doi: 10.1007/s00709-018-1280-7. Epub 2018 Jun 26. Protoplasma. 2019. PMID: 29946904
-
Functional Exploration of Chaperonin (HSP60/10) Family Genes and their Abiotic Stress-induced Expression Patterns in Sorghum bicolor.Curr Genomics. 2021 Feb;22(2):137-152. doi: 10.2174/1389202922666210324154336. Curr Genomics. 2021. PMID: 34220300 Free PMC article.
-
Transcriptomic analysis of the response of Avena sativa to Bacillus amyloliquefaciens DGL1.Front Microbiol. 2024 Apr 3;15:1321989. doi: 10.3389/fmicb.2024.1321989. eCollection 2024. Front Microbiol. 2024. PMID: 38633698 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

