Use of multiple time points to model parotid differentiation
- PMID: 26484231
- PMCID: PMC4583626
- DOI: 10.1016/j.gdata.2015.05.005
Use of multiple time points to model parotid differentiation
Abstract
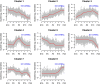
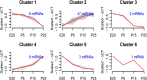
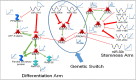
In order to understand the process of terminal differentiation in salivary acinar cells, mRNA and microRNA expression was measured across the month long process of differentiation in the parotid gland of the rat. Acinar cells were isolated at either nine time points (mRNA) or four time points (microRNA) in triplicate using laser capture microdissection (LCM). One of the values of this dataset comes from the high quality RNA (RIN > 7) that was used in this study, which can be prohibitively difficult to obtain from such an RNaseI-rich tissue. Global mRNA expression was measured by rat genome microarray hybridization (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65586), and expression of microRNAs by qPCR array (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE65324). Comparing expression at different ages, 2656 mRNAs and 64 microRNAs were identified as differentially expressed. Because mRNA expression was sampled at many time points, clustering and regression analysis were able to identify dynamic expression patterns that had not been implicated in acinar differentiation before. Integration of the two datasets allowed the identification of microRNA target genes, and a gene regulatory network. Bioinformatics R code and additional details of experimental methods and data analysis are provided.
Keywords: Differentiation; MicroRNAs; Microarray; Network analysis; Salivary gland.
Figures



Similar articles
-
A systems biology approach identifies a regulatory network in parotid acinar cell terminal differentiation.PLoS One. 2015 Apr 30;10(4):e0125153. doi: 10.1371/journal.pone.0125153. eCollection 2015. PLoS One. 2015. PMID: 25928148 Free PMC article.
-
Technical data of the transcriptomic analysis performed on tsetse fly symbionts, Sodalis glossinidius and Wigglesworthia glossinidia, harbored, respectively by non-infected, Trypanosoma brucei gambiense infected and self-cured Glossina palpalis gambiensis tsetse flies.Genom Data. 2015 Apr 14;4:133-6. doi: 10.1016/j.gdata.2015.04.007. eCollection 2015 Jun. Genom Data. 2015. PMID: 26484198 Free PMC article.
-
Mapping transcriptome profiles of in vitro iPSC-derived cardiac differentiation to in utero heart development.Genom Data. 2015 Dec 30;7:129-30. doi: 10.1016/j.gdata.2015.12.022. eCollection 2016 Mar. Genom Data. 2015. PMID: 26981387 Free PMC article.
-
MicroRNA expression profiles of trophoblastic cells.Placenta. 2012 Sep;33(9):725-34. doi: 10.1016/j.placenta.2012.05.009. Epub 2012 Jun 19. Placenta. 2012. PMID: 22721760
-
Retrospective analysis: reproducibility of interblastomere differences of mRNA expression in 2-cell stage mouse embryos is remarkably poor due to combinatorial mechanisms of blastomere diversification.Mol Hum Reprod. 2018 Jul 1;24(7):388-400. doi: 10.1093/molehr/gay021. Mol Hum Reprod. 2018. PMID: 29746690
References
-
- Sivakumar S., Mirels L., Miranda A.J., Hand A.R. Secretory protein expression patterns during rat parotid gland development. Anat. Rec. 1998;252(3):485–497. - PubMed
-
- Redman R.S., Sreebny L.M. Morphologic and biochemical observations on the development of the rat parotid gland. Dev. Biol. 1971;25(2):248–279. - PubMed
-
- Gautier L., Cope L., Bolstad B.M., Irizarry R.A. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. - PubMed
-
- Gentleman R., Carey V., Hahne F. 2015. Genefilter: methods for filtering genes from high throughput experiments, R package version 1.50.0.
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

