Crystal Structure of the Human Cytomegalovirus Glycoprotein B
- PMID: 26484870
- PMCID: PMC4617298
- DOI: 10.1371/journal.ppat.1005227
Crystal Structure of the Human Cytomegalovirus Glycoprotein B
Erratum in
-
Correction: Crystal Structure of the Human Cytomegalovirus Glycoprotein B.PLoS Pathog. 2015 Nov 17;11(11):e1005300. doi: 10.1371/journal.ppat.1005300. eCollection 2015 Nov. PLoS Pathog. 2015. PMID: 26575647 Free PMC article. No abstract available.
Abstract
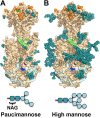
Human cytomegalovirus (HCMV), a dsDNA, enveloped virus, is a ubiquitous pathogen that establishes lifelong latent infections and caused disease in persons with compromised immune systems, e.g., organ transplant recipients or AIDS patients. HCMV is also a leading cause of congenital viral infections in newborns. Entry of HCMV into cells requires the conserved glycoprotein B (gB), thought to function as a fusogen and reported to bind signaling receptors. gB also elicits a strong immune response in humans and induces the production of neutralizing antibodies although most anti-gB Abs are non-neutralizing. Here, we report the crystal structure of the HCMV gB ectodomain determined to 3.6-Å resolution, which is the first atomic-level structure of any betaherpesvirus glycoprotein. The structure of HCMV gB resembles the postfusion structures of HSV-1 and EBV homologs, establishing it as a new member of the class III viral fusogens. Despite structural similarities, each gB has a unique domain arrangement, demonstrating structural plasticity of gB that may accommodate virus-specific functional requirements. The structure illustrates how extensive glycosylation of the gB ectodomain influences antibody recognition. Antigenic sites that elicit neutralizing antibodies are more heavily glycosylated than those that elicit non-neutralizing antibodies, which suggest that HCMV gB uses glycans to shield neutralizing epitopes while exposing non-neutralizing epitopes. This glycosylation pattern may have evolved to direct the immune response towards generation of non-neutralizing antibodies thus helping HCMV to avoid clearance. HCMV gB structure provides a starting point for elucidation of its antigenic and immunogenic properties and aid in the design of recombinant vaccines and monoclonal antibody therapies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody.Nat Commun. 2015 Sep 14;6:8176. doi: 10.1038/ncomms9176. Nat Commun. 2015. PMID: 26365435 Free PMC article.
-
Identification of a neutralizing epitope within antigenic domain 5 of glycoprotein B of human cytomegalovirus.J Virol. 2015 Jan;89(1):361-72. doi: 10.1128/JVI.02393-14. Epub 2014 Oct 15. J Virol. 2015. PMID: 25320309 Free PMC article.
-
Recognition of a highly conserved glycoprotein B epitope by a bivalent antibody neutralizing HCMV at a post-attachment step.PLoS Pathog. 2020 Aug 3;16(8):e1008736. doi: 10.1371/journal.ppat.1008736. eCollection 2020 Aug. PLoS Pathog. 2020. PMID: 32745149 Free PMC article.
-
Human cytomegalovirus (HCMV) infection/re-infection: development of a protective HCMV vaccine.New Microbiol. 2019 Jan;42(1):1-20. Epub 2019 Jan 21. New Microbiol. 2019. PMID: 30671581 Review.
-
The Humoral Immune Response Against the gB Vaccine: Lessons Learnt from Protection in Solid Organ Transplantation.Vaccines (Basel). 2019 Jul 17;7(3):67. doi: 10.3390/vaccines7030067. Vaccines (Basel). 2019. PMID: 31319553 Free PMC article. Review.
Cited by
-
The Glycoprotein B Cytoplasmic Domain Lysine Cluster Is Critical for Varicella-Zoster Virus Cell-Cell Fusion Regulation and Infection.J Virol. 2016 Dec 16;91(1):e01707-16. doi: 10.1128/JVI.01707-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795427 Free PMC article.
-
Pathogen at the Gates: Human Cytomegalovirus Entry and Cell Tropism.Viruses. 2018 Dec 11;10(12):704. doi: 10.3390/v10120704. Viruses. 2018. PMID: 30544948 Free PMC article. Review.
-
Expression, Purification, and Crystallization of Full-Length HSV-1 gB for Structure Determination.Methods Mol Biol. 2020;2060:395-407. doi: 10.1007/978-1-4939-9814-2_24. Methods Mol Biol. 2020. PMID: 31617193 Free PMC article.
-
The Structures and Functions of VZV Glycoproteins.Curr Top Microbiol Immunol. 2023;438:25-58. doi: 10.1007/82_2021_243. Curr Top Microbiol Immunol. 2023. PMID: 34731265 Free PMC article.
-
Association of Single-Nucleotide Polymorphisms in DC-SIGN with Nasopharyngeal Carcinoma Susceptibility.Dis Markers. 2017;2017:6309754. doi: 10.1155/2017/6309754. Epub 2017 Jun 14. Dis Markers. 2017. PMID: 28694559 Free PMC article.
References
-
- CDC. Congenital CMV Infection2010. Available from: http://www.cdc.gov/cmv/congenital-infection.html.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

