Energy Metabolism in Mesenchymal Stem Cells During Osteogenic Differentiation
- PMID: 26487485
- PMCID: PMC4733323
- DOI: 10.1089/scd.2015.0193
Energy Metabolism in Mesenchymal Stem Cells During Osteogenic Differentiation
Abstract
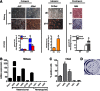
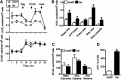
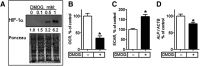
There is emerging interest in stem cell energy metabolism and its effect on differentiation. Bioenergetic changes in differentiating bone marrow mesenchymal stem cells (MSCs) are poorly understood and were the focus of our study. Using bioenergetic profiling and transcriptomics, we have established that MSCs activate the mitochondrial process of oxidative phosphorylation (OxPhos) during osteogenic differentiation, but they maintain levels of glycolysis similar to undifferentiated cells. Consistent with their glycolytic phenotype, undifferentiated MSCs have high levels of hypoxia-inducible factor 1 (HIF-1). Osteogenically induced MSCs downregulate HIF-1 and this downregulation is required for activation of OxPhos. In summary, our work provides important insights on MSC bioenergetics and proposes a HIF-based mechanism of regulation of mitochondrial OxPhos in MSCs.
Figures



References
-
- Manolagas SC. (2000). Birth and death of bone cells: basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr Rev 21:115–137 - PubMed
-
- Baker N, Boyette LB. and Tuan RS. (2015). Characterization of bone marrow-derived mesenchymal stem cells in aging. Bone 70:37–47 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

