Agonist antibody that induces human malignant cells to kill one another
- PMID: 26487683
- PMCID: PMC4653151
- DOI: 10.1073/pnas.1519079112
Agonist antibody that induces human malignant cells to kill one another
Abstract
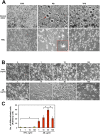
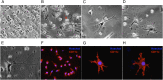
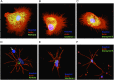
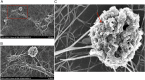
An attractive, but as yet generally unrealized, approach to cancer therapy concerns discovering agents that change the state of differentiation of the cancer cells. Recently, we discovered a phenomenon that we call "receptor pleiotropism" in which agonist antibodies against known receptors induce cell fates that are very different from those induced by the natural agonist to the same receptor. Here, we show that one can take advantage of this phenomenon to convert acute myeloblastic leukemic cells into natural killer cells. Upon induction with the antibody, these leukemic cells enter into a differentiation cascade in which as many as 80% of the starting leukemic cells can be differentiated. The antibody-induced killer cells make large amounts of perforin, IFN-γ, and granzyme B and attack and kill other members of the leukemic cell population. Importantly, induction of killer cells is confined to transformed cells, in that normal bone marrow cells are not induced to form killer cells. Thus, it seems possible to use agonist antibodies to change the differentiation state of cancer cells into those that attack and kill other members of the malignant clone from which they originate.
Keywords: agonist antibody; combinatorial antibody libraries; differentiation; natural killer cell.
Conflict of interest statement
Conflict of interest statement: R.A.L. is a founder of Zebra biologics.
Figures
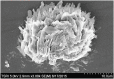
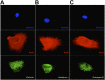
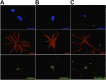
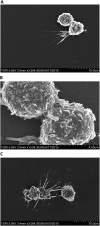
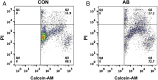
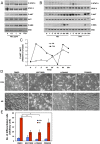
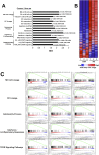

Comment in
-
Activating pleiotropic receptors to kill cancer cells.Cell Cycle. 2016;15(2):158-9. doi: 10.1080/15384101.2015.1118912. Epub 2015 Nov 20. Cell Cycle. 2016. PMID: 26588245 Free PMC article. No abstract available.
Similar articles
-
An agonist antibody prefers relapsed AML for induction of cells that kill each other.Sci Rep. 2019 Mar 5;9(1):3494. doi: 10.1038/s41598-019-40087-7. Sci Rep. 2019. PMID: 30837591 Free PMC article.
-
Activating pleiotropic receptors to kill cancer cells.Cell Cycle. 2016;15(2):158-9. doi: 10.1080/15384101.2015.1118912. Epub 2015 Nov 20. Cell Cycle. 2016. PMID: 26588245 Free PMC article. No abstract available.
-
[Changes of natural kill cell in peripheral blood of patients with myelodysplastic syndrome].Zhonghua Yi Xue Za Zhi. 2014 Mar 18;94(10):737-41. Zhonghua Yi Xue Za Zhi. 2014. PMID: 24844955 Chinese.
-
[AML stem cell].Rinsho Ketsueki. 2013 Oct;54(10):1643-50. Rinsho Ketsueki. 2013. PMID: 24064813 Review. Japanese. No abstract available.
-
Expression of multiple granzymes by cytotoxic T lymphocyte implies that they activate diverse apoptotic pathways in target cells.Int Rev Immunol. 2010;29(1):38-55. doi: 10.3109/08830180903247889. Int Rev Immunol. 2010. PMID: 20100081 Review.
Cited by
-
Selection of a Full Agonist Combinatorial Antibody that Rescues Leptin Deficiency In Vivo.Adv Sci (Weinh). 2020 Jul 1;7(16):2000818. doi: 10.1002/advs.202000818. eCollection 2020 Aug. Adv Sci (Weinh). 2020. PMID: 32832353 Free PMC article.
-
Antibody Libraries as Tools to Discover Functional Antibodies and Receptor Pleiotropism.Int J Mol Sci. 2021 Apr 16;22(8):4123. doi: 10.3390/ijms22084123. Int J Mol Sci. 2021. PMID: 33923551 Free PMC article. Review.
-
Target neutrophil heterogeneity and plasticity in cancer.J Hematol Oncol. 2025 Aug 12;18(1):79. doi: 10.1186/s13045-025-01731-0. J Hematol Oncol. 2025. PMID: 40797279 Free PMC article. Review.
-
Migration-based selections of antibodies that convert bone marrow into trafficking microglia-like cells that reduce brain amyloid β.Proc Natl Acad Sci U S A. 2018 Jan 16;115(3):E372-E381. doi: 10.1073/pnas.1719259115. Epub 2018 Jan 2. Proc Natl Acad Sci U S A. 2018. PMID: 29295920 Free PMC article.
-
Emerging principles of cytokine pharmacology and therapeutics.Nat Rev Drug Discov. 2023 Jan;22(1):21-37. doi: 10.1038/s41573-022-00557-6. Epub 2022 Sep 21. Nat Rev Drug Discov. 2023. PMID: 36131080 Free PMC article. Review.
References
-
- Kershaw MH, Westwood JA, Darcy PK. Gene-engineered T cells for cancer therapy. Nat Rev Cancer. 2013;13(8):525–541. - PubMed
-
- Waldhauer I, Steinle A. NK cells and cancer immunosurveillance. Oncogene. 2008;27(45):5932–5943. - PubMed
-
- Lanier LL. NK cell receptors. Annu Rev Immunol. 1998;16:359–393. - PubMed
-
- Childs RW, Carlsten M. Therapeutic approaches to enhance natural killer cell cytotoxicity against cancer: The force awakens. Nat Rev Drug Discov. 2015;14(7):487–498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

