Defining the Apoptotic Trigger: THE INTERACTION OF CYTOCHROME c AND CARDIOLIPIN
- PMID: 26487716
- PMCID: PMC4692216
- DOI: 10.1074/jbc.M115.689406
Defining the Apoptotic Trigger: THE INTERACTION OF CYTOCHROME c AND CARDIOLIPIN
Abstract
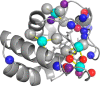
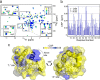
The interaction between cytochrome c and the anionic lipid cardiolipin has been proposed as a primary event in the apoptotic signaling cascade. Numerous studies that have examined the interaction of cytochrome c with cardiolipin embedded in a variety of model phospholipid membranes have suggested that partial unfolding of the protein is a precursor to the apoptotic response. However, these studies lacked site resolution and used model systems with negligible or a positive membrane curvature, which is distinct from the large negative curvature of the invaginations of the inner mitochondrial membrane where cytochrome c resides. We have used reverse micelle encapsulation to mimic the potential effects of confinement on the interaction of cytochrome c with cardiolipin. Encapsulation of oxidized horse cytochrome c in 1-decanoyl-rac-glycerol/lauryldimethylamine-N-oxide/hexanol reverse micelles prepared in pentane yields NMR spectra essentially identical to the protein in free aqueous solution. The structure of encapsulated ferricytochrome c was determined to high precision (<r.m.s. deviation>bb ∼ 0.23 Å) using NMR-based methods and is closely similar to the cryogenic crystal structure (<r.m.s. deviation>bb ∼ 1.2 Å). Incorporation of cardiolipin into the reverse micelle surfactant shell causes localized chemical shift perturbations of the encapsulated protein, providing the first view of the cardiolipin/cytochrome c interaction interface at atomic resolution. Three distinct sites of interaction are detected: the so-called A- and L-sites, plus a previously undocumented interaction centered on residues Phe-36, Gly-37, Thr-58, Trp-59, and Lys-60. Importantly, in distinct contrast to earlier studies of this interaction, the protein is not significantly disturbed by the binding of cardiolipin in the context of the reverse micelle.
Keywords: apoptosis; cardiolipin; confined space; cytochrome c; lipid-protein interaction; nuclear magnetic resonance (NMR); protein folding; protein hydration; protein stability.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures
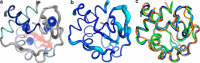

References
-
- Vaux D. L. (2011) Apoptogenic factors released from mitochondria. Biochim. Biophys. Acta 1813, 546–550 - PubMed
-
- Kagan V. E., Bayir H. A., Belikova N. A., Kapralov O., Tyurina Y. Y., Tyurin V. A., Jiang J., Stoyanovsky D. A., Wipf P., Kochanek P. M., Greenberger J. S., Pitt B., Shvedova A. A., and Borisenko G. (2009) Cytochrome c/cardiolipin relations in mitochondria: a kiss of death. Free Radic. Biol. Med. 46, 1439–1453 - PMC - PubMed
-
- Rytömaa M., and Kinnunen P. K. (1994) Evidence for two distinct acidic phospholipid-binding sites in cytochrome c. J. Biol. Chem. 269, 1770–1774 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

