Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea
- PMID: 26487780
- PMCID: PMC4631768
- DOI: 10.1242/dev.126763
Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea
Erratum in
-
Epigenetic regulation of Atoh1 guides hair cell development in the mammalian cochlea.Development. 2016 May 1;143(9):1632. doi: 10.1242/dev.137976. Development. 2016. PMID: 27143757 Free PMC article. No abstract available.
Abstract
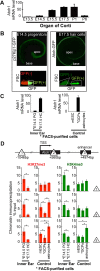
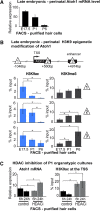
In the developing cochlea, sensory hair cell differentiation depends on the regulated expression of the bHLH transcription factor Atoh1. In mammals, if hair cells die they do not regenerate, leading to permanent deafness. By contrast, in non-mammalian vertebrates robust regeneration occurs through upregulation of Atoh1 in the surviving supporting cells that surround hair cells, leading to functional recovery. Investigation of crucial transcriptional events in the developing organ of Corti, including those involving Atoh1, has been hampered by limited accessibility to purified populations of the small number of cells present in the inner ear. We used µChIP and qPCR assays of FACS-purified cells to track changes in the epigenetic status of the Atoh1 locus during sensory epithelia development in the mouse. Dynamic changes in the histone modifications H3K4me3/H3K27me3, H3K9ac and H3K9me3 reveal a progression from poised, to active, to repressive marks, correlating with the onset of Atoh1 expression and its subsequent silencing during the perinatal (P1 to P6) period. Inhibition of acetylation blocked the increase in Atoh1 mRNA in nascent hair cells, as well as ongoing hair cell differentiation during embryonic organ of Corti development ex vivo. These results reveal an epigenetic mechanism of Atoh1 regulation underlying hair cell differentiation and subsequent maturation. Interestingly, the H3K4me3/H3K27me3 bivalent chromatin structure observed in progenitors persists at the Atoh1 locus in perinatal supporting cells, suggesting an explanation for the latent capacity of these cells to transdifferentiate into hair cells, and highlighting their potential as therapeutic targets in hair cell regeneration.
Keywords: Epigenetics of Atoh1 regulation; Epigenetics of inner ear development; Mouse; Sensory hair cell differentiation.
© 2015. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures


Similar articles
-
Selection of cell fate in the organ of Corti involves the integration of Hes/Hey signaling at the Atoh1 promoter.Development. 2016 Mar 1;143(5):841-50. doi: 10.1242/dev.129320. Development. 2016. PMID: 26932672 Free PMC article.
-
A novel Atoh1 "self-terminating" mouse model reveals the necessity of proper Atoh1 level and duration for hair cell differentiation and viability.PLoS One. 2012;7(1):e30358. doi: 10.1371/journal.pone.0030358. Epub 2012 Jan 18. PLoS One. 2012. PMID: 22279587 Free PMC article.
-
Three-dimensional live imaging of Atoh1 reveals the dynamics of hair cell induction and organization in the developing cochlea.Development. 2019 Nov 1;146(21):dev177881. doi: 10.1242/dev.177881. Development. 2019. PMID: 31676552
-
Atoh1 and other related key regulators in the development of auditory sensory epithelium in the mammalian inner ear: function and interplay.Dev Biol. 2019 Feb 15;446(2):133-141. doi: 10.1016/j.ydbio.2018.12.025. Epub 2018 Dec 31. Dev Biol. 2019. PMID: 30605626 Review.
-
Beyond generalized hair cells: molecular cues for hair cell types.Hear Res. 2013 Mar;297:30-41. doi: 10.1016/j.heares.2012.11.008. Epub 2012 Nov 27. Hear Res. 2013. PMID: 23201032 Free PMC article. Review.
Cited by
-
Heat Stress Responsive Aux/IAA Protein, OsIAA29 Regulates Grain Filling Through OsARF17 Mediated Auxin Signaling Pathway.Rice (N Y). 2024 Feb 19;17(1):16. doi: 10.1186/s12284-024-00694-z. Rice (N Y). 2024. PMID: 38374238 Free PMC article.
-
Approaches for the study of epigenetic modifications in the inner ear and related tissues.Hear Res. 2019 May;376:69-85. doi: 10.1016/j.heares.2019.01.007. Epub 2019 Jan 12. Hear Res. 2019. PMID: 30679030 Free PMC article. Review.
-
Cell tropism of adeno-associated viruses within the mouse inner ear in vivo: from embryonic to adult stages.Sci Rep. 2025 Apr 18;15(1):13479. doi: 10.1038/s41598-025-98007-x. Sci Rep. 2025. PMID: 40251388 Free PMC article.
-
Insights into inner ear-specific gene regulation: Epigenetics and non-coding RNAs in inner ear development and regeneration.Semin Cell Dev Biol. 2017 May;65:69-79. doi: 10.1016/j.semcdb.2016.11.002. Epub 2016 Nov 9. Semin Cell Dev Biol. 2017. PMID: 27836639 Free PMC article. Review.
-
Sox2 interacts with Atoh1 and Huwe1 loci to regulate Atoh1 transcription and stability during hair cell differentiation.PLoS Genet. 2025 Jan 30;21(1):e1011573. doi: 10.1371/journal.pgen.1011573. eCollection 2025 Jan. PLoS Genet. 2025. PMID: 39883720 Free PMC article.
References
-
- Akazawa C., Ishibashi M., Shimizu C., Nakanishi S. and Kageyama R. (1995). A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J. Biol. Chem. 270, 8730-8738. 10.1074/jbc.270.15.8730 - DOI - PubMed
-
- Balasubramanyam K., Varier R. A., Altaf M., Swaminathan V., Siddappa N. B., Ranga U. and Kundu T. K. (2004). Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279, 51163-51171. 10.1074/jbc.M409024200 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

