The Low Noise Limit in Gene Expression
- PMID: 26488303
- PMCID: PMC4619080
- DOI: 10.1371/journal.pone.0140969
The Low Noise Limit in Gene Expression
Abstract
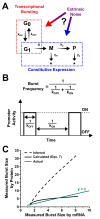
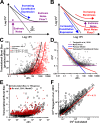
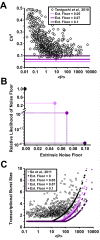
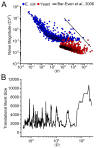
Protein noise measurements are increasingly used to elucidate biophysical parameters. Unfortunately noise analyses are often at odds with directly measured parameters. Here we show that these inconsistencies arise from two problematic analytical choices: (i) the assumption that protein translation rate is invariant for different proteins of different abundances, which has inadvertently led to (ii) the assumption that a large constitutive extrinsic noise sets the low noise limit in gene expression. While growing evidence suggests that transcriptional bursting may set the low noise limit, variability in translational bursting has been largely ignored. We show that genome-wide systematic variation in translational efficiency can-and in the case of E. coli does-control the low noise limit in gene expression. Therefore constitutive extrinsic noise is small and only plays a role in the absence of a systematic variation in translational efficiency. These results show the existence of two distinct expression noise patterns: (1) a global noise floor uniformly imposed on all genes by expression bursting; and (2) high noise distributed to only a select group of genes.
Conflict of interest statement
Figures


Similar articles
-
Transcript level and sequence determinants of protein abundance and noise in Escherichia coli.Nucleic Acids Res. 2014 Apr;42(8):4791-9. doi: 10.1093/nar/gku126. Epub 2014 Feb 7. Nucleic Acids Res. 2014. PMID: 24510099 Free PMC article.
-
Experimental measurements and mathematical modeling of biological noise arising from transcriptional and translational regulation of basic synthetic gene circuits.J Theor Biol. 2016 Apr 21;395:153-160. doi: 10.1016/j.jtbi.2016.02.004. Epub 2016 Feb 10. J Theor Biol. 2016. PMID: 26874228
-
Codon influence on protein expression in E. coli correlates with mRNA levels.Nature. 2016 Jan 21;529(7586):358-363. doi: 10.1038/nature16509. Epub 2016 Jan 13. Nature. 2016. PMID: 26760206 Free PMC article.
-
Optimizing scaleup yield for protein production: Computationally Optimized DNA Assembly (CODA) and Translation Engineering.Biotechnol Annu Rev. 2007;13:27-42. doi: 10.1016/S1387-2656(07)13002-7. Biotechnol Annu Rev. 2007. PMID: 17875472 Review.
-
Noise in bacterial gene expression.Biochem Soc Trans. 2019 Feb 28;47(1):209-217. doi: 10.1042/BST20180500. Epub 2018 Dec 21. Biochem Soc Trans. 2019. PMID: 30578346 Review.
Cited by
-
Cell Size-Based Decision-Making of a Viral Gene Circuit.Cell Rep. 2018 Dec 26;25(13):3844-3857.e5. doi: 10.1016/j.celrep.2018.12.009. Cell Rep. 2018. PMID: 30590053 Free PMC article.
-
Optimisation of gene expression noise for cellular persistence against lethal events.J Theor Biol. 2025 Feb 7;598:111996. doi: 10.1016/j.jtbi.2024.111996. Epub 2024 Nov 25. J Theor Biol. 2025. PMID: 39603338
-
Homeostasis of protein and mRNA concentrations in growing cells.Nat Commun. 2018 Oct 29;9(1):4496. doi: 10.1038/s41467-018-06714-z. Nat Commun. 2018. PMID: 30374016 Free PMC article.
-
The Pathway To Establishing HIV Latency Is Critical to How Latency Is Maintained and Reversed.J Virol. 2018 Jun 13;92(13):e02225-17. doi: 10.1128/JVI.02225-17. Print 2018 Jul 1. J Virol. 2018. PMID: 29643247 Free PMC article.
-
N-terminal tagging of RNA Polymerase II shapes transcriptomes more than C-terminal alterations.iScience. 2024 May 7;27(6):109914. doi: 10.1016/j.isci.2024.109914. eCollection 2024 Jun 21. iScience. 2024. PMID: 38799575 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

