Digital signaling decouples activation probability and population heterogeneity
- PMID: 26488364
- PMCID: PMC4608393
- DOI: 10.7554/eLife.08931
Digital signaling decouples activation probability and population heterogeneity
Abstract
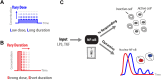
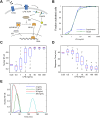
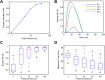
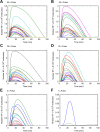
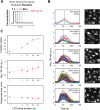
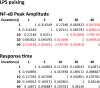
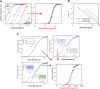
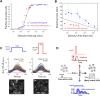
Digital signaling enhances robustness of cellular decisions in noisy environments, but it is unclear how digital systems transmit temporal information about a stimulus. To understand how temporal input information is encoded and decoded by the NF-κB system, we studied transcription factor dynamics and gene regulation under dose- and duration-modulated inflammatory inputs. Mathematical modeling predicted and microfluidic single-cell experiments confirmed that integral of the stimulus (or area, concentration × duration) controls the fraction of cells that activate NF-κB in the population. However, stimulus temporal profile determined NF-κB dynamics, cell-to-cell variability, and gene expression phenotype. A sustained, weak stimulation lead to heterogeneous activation and delayed timing that is transmitted to gene expression. In contrast, a transient, strong stimulus with the same area caused rapid and uniform dynamics. These results show that digital NF-κB signaling enables multidimensional control of cellular phenotype via input profile, allowing parallel and independent control of single-cell activation probability and population heterogeneity.
Keywords: cell-to-cell heterogeneity; computational biology; digital signaling; immunology; innate immunity; mouse; signaling dynamics; single-cell analysis; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures










Similar articles
-
Processing stimulus dynamics by the NF-κB network in single cells.Exp Mol Med. 2023 Dec;55(12):2531-2540. doi: 10.1038/s12276-023-01133-7. Epub 2023 Dec 1. Exp Mol Med. 2023. PMID: 38040923 Free PMC article. Review.
-
Noise facilitates transcriptional control under dynamic inputs.Cell. 2015 Jan 29;160(3):381-92. doi: 10.1016/j.cell.2015.01.013. Cell. 2015. PMID: 25635454
-
NF-κB memory coordinates transcriptional responses to dynamic inflammatory stimuli.Cell Rep. 2022 Aug 16;40(7):111159. doi: 10.1016/j.celrep.2022.111159. Cell Rep. 2022. PMID: 35977475 Free PMC article.
-
Single-cell analyses reveal an attenuated NF-κB response in the Salmonella-infected fibroblast.Virulence. 2017 Aug 18;8(6):719-740. doi: 10.1080/21505594.2016.1229727. Epub 2016 Aug 30. Virulence. 2017. PMID: 27575017 Free PMC article.
-
Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP).Cell Signal. 2012 Apr;24(4):835-45. doi: 10.1016/j.cellsig.2011.12.006. Epub 2011 Dec 11. Cell Signal. 2012. PMID: 22182507 Review.
Cited by
-
Multiplexed microfluidic chip for cell co-culture.Analyst. 2022 Nov 21;147(23):5409-5418. doi: 10.1039/d2an01344d. Analyst. 2022. PMID: 36300548 Free PMC article.
-
Processing stimulus dynamics by the NF-κB network in single cells.Exp Mol Med. 2023 Dec;55(12):2531-2540. doi: 10.1038/s12276-023-01133-7. Epub 2023 Dec 1. Exp Mol Med. 2023. PMID: 38040923 Free PMC article. Review.
-
Integrating Immunology and Microfluidics for Single Immune Cell Analysis.Front Immunol. 2018 Oct 16;9:2373. doi: 10.3389/fimmu.2018.02373. eCollection 2018. Front Immunol. 2018. PMID: 30459757 Free PMC article. Review.
-
Cellular Decision Making by Non-Integrative Processing of TLR Inputs.Cell Rep. 2017 Apr 4;19(1):125-135. doi: 10.1016/j.celrep.2017.03.027. Cell Rep. 2017. PMID: 28380352 Free PMC article.
-
Adequate immune response ensured by binary IL-2 and graded CD25 expression in a murine transfer model.Elife. 2016 Dec 30;5:e20616. doi: 10.7554/eLife.20616. Elife. 2016. PMID: 28035902 Free PMC article.
References
-
- Ashall L, Horton CA, Nelson DE, Paszek P, Harper CV, Sillitoe K, Ryan S, Spiller DG, Unitt JF, Broomhead DS, Kell DB, Rand DA, Sée V, White MR. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

