A Conserved Secondary Structural Element in the Coding Region of the Influenza A Virus Nucleoprotein (NP) mRNA Is Important for the Regulation of Viral Proliferation
- PMID: 26488402
- PMCID: PMC4619443
- DOI: 10.1371/journal.pone.0141132
A Conserved Secondary Structural Element in the Coding Region of the Influenza A Virus Nucleoprotein (NP) mRNA Is Important for the Regulation of Viral Proliferation
Abstract
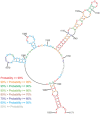
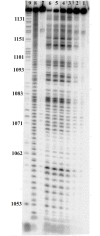
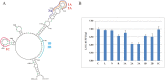
Influenza A virus is a threat to humans due to seasonal epidemics and infrequent, but dangerous, pandemics that lead to widespread infection and death. Eight segments of RNA constitute the genome of this virus and they encode greater than eight proteins via alternative splicing of coding (+)RNAs generated from the genomic (-)RNA template strand. RNA is essential in its life cycle. A bioinformatics analysis of segment 5, which encodes nucleoprotein, revealed a conserved structural motif in the (+)RNA. The secondary structure proposed by energy minimization and comparative analysis agrees with structure predicted based on experimental data using a 121 nucleotide in vitro RNA construct comprising an influenza A virus consensus sequence and also an entire segment 5 (+)RNA (strain A/VietNam/1203/2004 (H5N1)). The conserved motif consists of three hairpins with one being especially thermodynamically stable. The biological importance of this conserved secondary structure is supported in experiments using antisense oligonucleotides in cell line, which found that disruption of this motif led to inhibition of viral fitness. These results suggest that this conserved motif in the segment 5 (+)RNA might be a candidate for oligonucleotide-based antiviral therapy.
Conflict of interest statement
Figures



Similar articles
-
Influenza virus segment 5 (+)RNA - secondary structure and new targets for antiviral strategies.Sci Rep. 2017 Nov 8;7(1):15041. doi: 10.1038/s41598-017-15317-5. Sci Rep. 2017. PMID: 29118447 Free PMC article.
-
Construction of influenza virus siRNA expression vectors and their inhibitory effects on multiplication of influenza virus.Avian Dis. 2005 Dec;49(4):562-73. doi: 10.1637/7365-041205R2.1. Avian Dis. 2005. PMID: 16405000
-
Secondary Structure of Influenza A Virus Genomic Segment 8 RNA Folded in a Cellular Environment.Int J Mol Sci. 2022 Feb 23;23(5):2452. doi: 10.3390/ijms23052452. Int J Mol Sci. 2022. PMID: 35269600 Free PMC article.
-
Structure and assembly of the influenza A virus ribonucleoprotein complex.FEBS Lett. 2013 Apr 17;587(8):1206-14. doi: 10.1016/j.febslet.2013.02.048. Epub 2013 Mar 13. FEBS Lett. 2013. PMID: 23499938 Review.
-
[Classification and genome structure of influenza virus].Nihon Rinsho. 2003 Nov;61(11):1886-91. Nihon Rinsho. 2003. PMID: 14619426 Review. Japanese.
Cited by
-
The effective rate of influenza reassortment is limited during human infection.PLoS Pathog. 2017 Feb 7;13(2):e1006203. doi: 10.1371/journal.ppat.1006203. eCollection 2017 Feb. PLoS Pathog. 2017. PMID: 28170438 Free PMC article.
-
In vivo analysis of influenza A mRNA secondary structures identifies critical regulatory motifs.Nucleic Acids Res. 2019 Jul 26;47(13):7003-7017. doi: 10.1093/nar/gkz318. Nucleic Acids Res. 2019. PMID: 31053845 Free PMC article.
-
RNA2DMut: a web tool for the design and analysis of RNA structure mutations.RNA. 2018 Mar;24(3):273-286. doi: 10.1261/rna.063933.117. Epub 2017 Nov 28. RNA. 2018. PMID: 29183923 Free PMC article.
-
Self-Folding of Naked Segment 8 Genomic RNA of Influenza A Virus.PLoS One. 2016 Feb 5;11(2):e0148281. doi: 10.1371/journal.pone.0148281. eCollection 2016. PLoS One. 2016. PMID: 26848969 Free PMC article.
-
The effects of structure on pre-mRNA processing and stability.Methods. 2017 Aug 1;125:36-44. doi: 10.1016/j.ymeth.2017.06.001. Epub 2017 Jun 6. Methods. 2017. PMID: 28595983 Free PMC article. Review.
References
-
- Shapshak P, Chiappelli F, Somboonwit C, Sinnott J. The Influenza Pandemic of 2009 Lessons and Implications. Molecular Diagnosis & Therapy. 2011. 2011;15(2):63–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

