EAST Organizes Drosophila Insulator Proteins in the Interchromosomal Nuclear Compartment and Modulates CP190 Binding to Chromatin
- PMID: 26489095
- PMCID: PMC4638101
- DOI: 10.1371/journal.pone.0140991
EAST Organizes Drosophila Insulator Proteins in the Interchromosomal Nuclear Compartment and Modulates CP190 Binding to Chromatin
Abstract
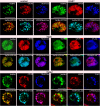
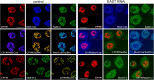
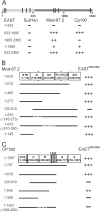
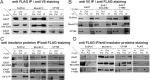
Recent data suggest that insulators organize chromatin architecture in the nucleus. The best studied Drosophila insulator proteins, dCTCF (a homolog of the vertebrate insulator protein CTCF) and Su(Hw), are DNA-binding zinc finger proteins. Different isoforms of the BTB-containing protein Mod(mdg4) interact with Su(Hw) and dCTCF. The CP190 protein is a cofactor for the dCTCF and Su(Hw) insulators. CP190 is required for the functional activity of insulator proteins and is involved in the aggregation of the insulator proteins into specific structures named nuclear speckles. Here, we have shown that the nuclear distribution of CP190 is dependent on the level of EAST protein, an essential component of the interchromatin compartment. EAST interacts with CP190 and Mod(mdg4)-67.2 proteins in vitro and in vivo. Over-expression of EAST in S2 cells leads to an extrusion of the CP190 from the insulator bodies containing Su(Hw), Mod(mdg4)-67.2, and dCTCF. In consistent with the role of the insulator bodies in assembly of protein complexes, EAST over-expression led to a striking decrease of the CP190 binding with the dCTCF and Su(Hw) dependent insulators and promoters. These results suggest that EAST is involved in the regulation of CP190 nuclear localization.
Conflict of interest statement
Figures


Similar articles
-
Interactions between BTB domain of CP190 and two adjacent regions in Su(Hw) are required for the insulator complex formation.Chromosoma. 2018 Mar;127(1):59-71. doi: 10.1007/s00412-017-0645-6. Epub 2017 Sep 22. Chromosoma. 2018. PMID: 28939920
-
Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila.Mol Cell. 2007 Dec 14;28(5):761-72. doi: 10.1016/j.molcel.2007.09.024. Mol Cell. 2007. PMID: 18082602 Free PMC article.
-
SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation.J Cell Sci. 2012 Apr 15;125(Pt 8):2064-74. doi: 10.1242/jcs.100172. Epub 2012 Feb 28. J Cell Sci. 2012. PMID: 22375064
-
Functional sub-division of the Drosophila genome via chromatin looping: the emerging importance of CP190.Nucleus. 2013 Mar-Apr;4(2):115-22. doi: 10.4161/nucl.23389. Epub 2013 Jan 18. Nucleus. 2013. PMID: 23333867 Free PMC article. Review.
-
Genetic and molecular analysis of the gypsy chromatin insulator of Drosophila.Proc Natl Acad Sci U S A. 1996 Sep 3;93(18):9378-83. doi: 10.1073/pnas.93.18.9378. Proc Natl Acad Sci U S A. 1996. PMID: 8790337 Free PMC article. Review.
Cited by
-
Multiple interactions are involved in a highly specific association of the Mod(mdg4)-67.2 isoform with the Su(Hw) sites in Drosophila.Open Biol. 2017 Oct;7(10):170150. doi: 10.1098/rsob.170150. Open Biol. 2017. PMID: 29021216 Free PMC article.
-
Interactions between BTB domain of CP190 and two adjacent regions in Su(Hw) are required for the insulator complex formation.Chromosoma. 2018 Mar;127(1):59-71. doi: 10.1007/s00412-017-0645-6. Epub 2017 Sep 22. Chromosoma. 2018. PMID: 28939920
-
Insulator speckles associated with long-distance chromatin contacts.Biol Open. 2016 Sep 15;5(9):1266-74. doi: 10.1242/bio.019455. Biol Open. 2016. PMID: 27464669 Free PMC article.
-
Role of Su(Hw) zinc finger 10 and interaction with CP190 and Mod(mdg4) proteins in recruiting the Su(Hw) complex to chromatin sites in Drosophila.PLoS One. 2018 Feb 23;13(2):e0193497. doi: 10.1371/journal.pone.0193497. eCollection 2018. PLoS One. 2018. PMID: 29474480 Free PMC article.
-
The Functions and Mechanisms of Action of Insulators in the Genomes of Higher Eukaryotes.Acta Naturae. 2020 Oct-Dec;12(4):15-33. doi: 10.32607/actanaturae.11144. Acta Naturae. 2020. PMID: 33456975 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

