Transcriptomic characterization of fibrolamellar hepatocellular carcinoma
- PMID: 26489647
- PMCID: PMC4640752
- DOI: 10.1073/pnas.1424894112
Transcriptomic characterization of fibrolamellar hepatocellular carcinoma
Abstract
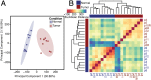
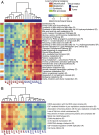
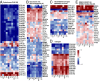
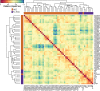
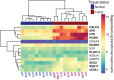
Fibrolamellar hepatocellular carcinoma (FLHCC) tumors all carry a deletion of ∼ 400 kb in chromosome 19, resulting in a fusion of the genes for the heat shock protein, DNAJ (Hsp40) homolog, subfamily B, member 1, DNAJB1, and the catalytic subunit of protein kinase A, PRKACA. The resulting chimeric transcript produces a fusion protein that retains kinase activity. No other recurrent genomic alterations have been identified. Here we characterize the molecular pathogenesis of FLHCC with transcriptome sequencing (RNA sequencing). Differential expression (tumor vs. adjacent normal tissue) was detected for more than 3,500 genes (log2 fold change ≥ 1, false discovery rate ≤ 0.01), many of which were distinct from those found in hepatocellular carcinoma. Expression of several known oncogenes, such as ErbB2 and Aurora Kinase A, was increased in tumor samples. These and other dysregulated genes may serve as potential targets for therapeutic intervention.
Keywords: fusion gene; genomics; liver cancer; pediatric cancer; protein kinase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Edmondson HA. Differential diagnosis of tumors and tumor-like lesions of liver in infancy and childhood. AMA J Dis Child. 1956;91(2):168–186. - PubMed
-
- Craig JR, Peters RL, Edmondson HA, Omata M. Fibrolamellar carcinoma of the liver: A tumor of adolescents and young adults with distinctive clinico-pathologic features. Cancer. 1980;46(2):372–379. - PubMed
-
- Katzenstein HM, et al. Fibrolamellar hepatocellular carcinoma in children and adolescents. Cancer. 2003;97(8):2006–2012. - PubMed
-
- Weeda VB, et al. Fibrolamellar variant of hepatocellular carcinoma does not have a better survival than conventional hepatocellular carcinoma--results and treatment recommendations from the Childhood Liver Tumour Strategy Group (SIOPEL) experience. Eur J Cancer. 2013;49(12):2698–2704. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

