Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells
- PMID: 26489895
- PMCID: PMC4649257
- DOI: 10.1016/j.stemcr.2015.09.009
Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells
Abstract
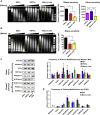
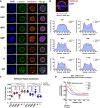
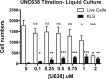
Epigenetic regulation serves as the basis for stem cell differentiation into distinct cell types, but it is unclear how global epigenetic changes are regulated during this process. Here, we tested the hypothesis that global chromatin organization affects the lineage potential of stem cells and that manipulation of chromatin dynamics influences stem cell function. Using nuclease sensitivity assays, we found a progressive decrease in chromatin digestion among pluripotent embryonic stem cells (ESCs), multipotent hematopoietic stem cells (HSCs), and mature hematopoietic cells. Quantitative high-resolution microscopy revealed that ESCs contain significantly more euchromatin than HSCs, with a further reduction in mature cells. Increased cellular maturation also led to heterochromatin localization to the nuclear periphery. Functionally, prevention of heterochromatin formation by inhibition of the histone methyltransferase G9A resulted in delayed HSC differentiation. Our results demonstrate global chromatin rearrangements during stem cell differentiation and that heterochromatin formation by H3K9 methylation regulates HSC differentiation.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Alabert C., Groth A. Chromatin replication and epigenome maintenance. Nat. Rev. Mol. Cell Biol. 2012;13:153–167. - PubMed
-
- Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A.G. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

