Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing
- PMID: 26490136
- PMCID: PMC4618736
- DOI: 10.1186/s12864-015-2010-6
Identification of novel and conserved microRNAs in Panax notoginseng roots by high-throughput sequencing
Abstract
Background: MicroRNAs (miRNAs) are small, non-coding RNAs that are important regulators of gene expression, and play major roles in plant development and their response to the environment. Root extracts from Panax notoginseng contain triterpene saponins as their principal bioactive constituent, and demonstrate medicinal properties. To investigate the novel and conserved miRNAs in P. notoginseng, three small RNA libraries constructed from 1-, 2-, and 3-year-old roots in which root saponin levels vary underwent high-throughput sequencing.
Methods: P. notoginseng roots, purified from 1-, 2-, and 3-year-old roots, were extracted for RNA, respectively. Three small libraries were constructed and subjected to next generation sequencing.
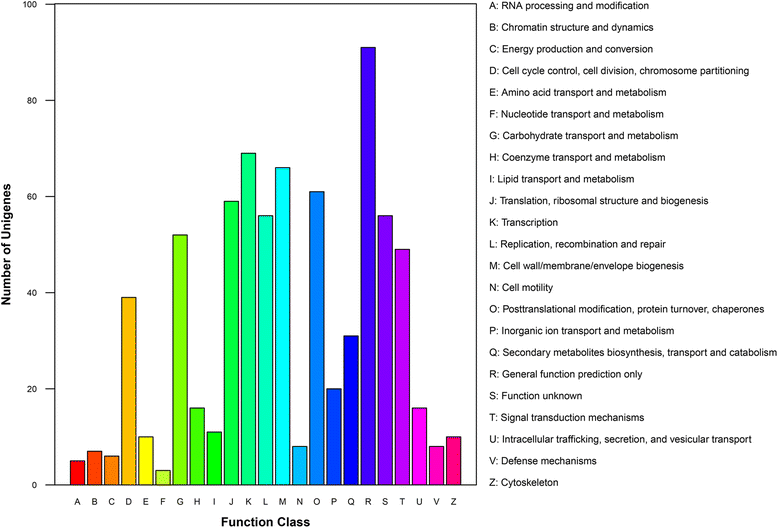
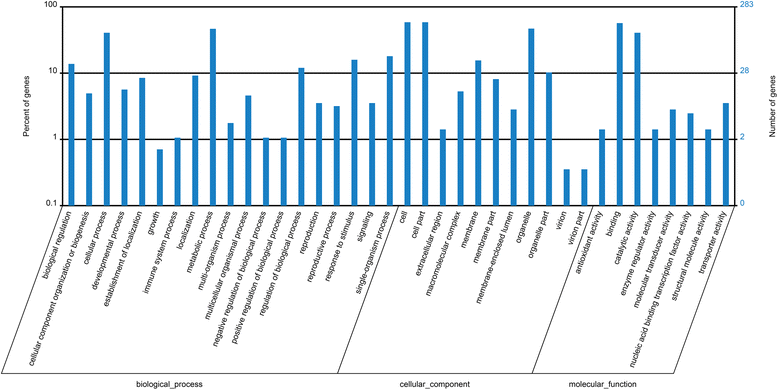
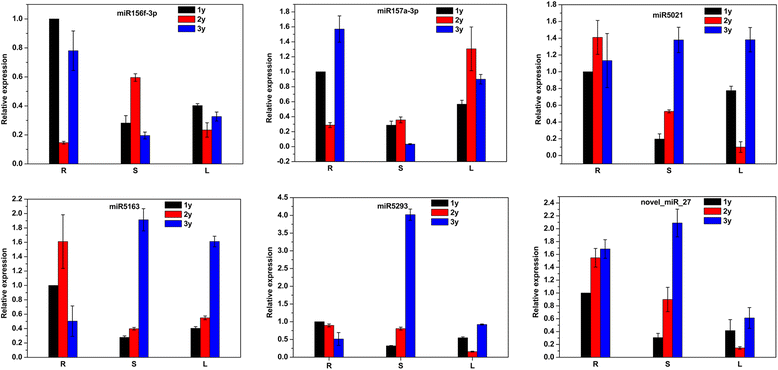
Results: Sequencing of the three libraries generated 67,217,124 clean reads from P. notoginseng roots. A total of 316 conserved miRNAs (belonging to 67 miRNA families and one unclassified family) and 52 novel miRNAs were identified. MIR156 and MIR166 were the largest miRNA families, while miR156i and miR156g showed the highest abundance of miRNA species. Potential miRNA target genes were predicted and annotated using Cluster of Orthologous Groups, Gene Ontology, and Kyoto Encyclopedia of Genes and Genomes. Comparing these miRNAs between root samples revealed 33 that were differentially expressed between 2- and 1-year-old roots (8 increased, 25 decreased), 27 differentially expressed between 3- and 1-year-old roots (7 increased, 20 decreased), and 29 differentially expressed between 3- and 2-year-old roots (8 increased, 21 decreased). Two significantly differentially expressed miRNAs and four miRNAs predicted to target genes involved in the terpenoid backbone biosynthesis pathway were selected and validated by quantitative reverse transcription PCR. Furthermore, the expression patterns of these six miRNAs were analyzed in P. notoginseng roots, stems, and leaves at different developmental stages.
Conclusions: This study identified a large number of P. notoginseng miRNAs and their target genes, functional annotations, and gene expression patterns. It provides the first known miRNA profiles of the P. notoginseng root development cycle.
Figures

Similar articles
-
Identification and expression profiling of Vigna mungo microRNAs from leaf small RNA transcriptome by deep sequencing.J Integr Plant Biol. 2014 Jan;56(1):15-23. doi: 10.1111/jipb.12115. Epub 2013 Dec 19. J Integr Plant Biol. 2014. PMID: 24138283
-
High-throughput sequencing and characterization of the small RNA transcriptome reveal features of novel and conserved microRNAs in Panax ginseng.PLoS One. 2012;7(9):e44385. doi: 10.1371/journal.pone.0044385. Epub 2012 Sep 4. PLoS One. 2012. PMID: 22962612 Free PMC article.
-
Small RNA profiles from Panax notoginseng roots differing in sizes reveal correlation between miR156 abundances and root biomass levels.Sci Rep. 2017 Aug 25;7(1):9418. doi: 10.1038/s41598-017-09670-8. Sci Rep. 2017. PMID: 28842680 Free PMC article.
-
Bioactive Proteins in Panax notoginseng Roots and Other Panax Species.Curr Protein Pept Sci. 2019;20(3):231-239. doi: 10.2174/1389203719666180612083650. Curr Protein Pept Sci. 2019. PMID: 29895241 Review.
-
Small but powerful: function of microRNAs in plant development.Plant Cell Rep. 2018 Mar;37(3):515-528. doi: 10.1007/s00299-017-2246-5. Epub 2018 Jan 9. Plant Cell Rep. 2018. PMID: 29318384 Review.
Cited by
-
A tissue-specific profile of miRNAs and their targets related to paeoniaflorin and monoterpenoids biosynthesis in Paeonia lactiflora Pall. by transcriptome, small RNAs and degradome sequencing.PLoS One. 2023 Jan 26;18(1):e0279992. doi: 10.1371/journal.pone.0279992. eCollection 2023. PLoS One. 2023. PMID: 36701382 Free PMC article.
-
Panax notoginseng saponins radiosensitize colorectal cancer cells by regulating the SNHG6/miR-137 axis.RSC Adv. 2019 Nov 26;9(66):38558-38567. doi: 10.1039/c9ra07622k. eCollection 2019 Nov 25. RSC Adv. 2019. PMID: 35540209 Free PMC article.
-
Integration of Transcriptomes, Small RNAs, and Degradome Sequencing to Identify Putative miRNAs and Their Targets Related to Eu-Rubber Biosynthesis in Eucommia ulmoides.Genes (Basel). 2019 Aug 19;10(8):623. doi: 10.3390/genes10080623. Genes (Basel). 2019. PMID: 31430866 Free PMC article.
-
Integrative analysis of microRNAs and mRNAs reveals the regulatory networks of triterpenoid saponin metabolism in Soapberry (Sapindus mukorossi Gaertn.).Front Plant Sci. 2023 Jan 9;13:1037784. doi: 10.3389/fpls.2022.1037784. eCollection 2022. Front Plant Sci. 2023. PMID: 36699854 Free PMC article.
-
De novo assembly and annotation of the Zhe-Maidong (Ophiopogon japonicus (L.f.) Ker-Gawl) transcriptome in different growth stages.Sci Rep. 2017 Jun 15;7(1):3616. doi: 10.1038/s41598-017-03937-w. Sci Rep. 2017. PMID: 28620183 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

