Modeling of autosomal-dominant retinitis pigmentosa in Caenorhabditis elegans uncovers a nexus between global impaired functioning of certain splicing factors and cell type-specific apoptosis
- PMID: 26490224
- PMCID: PMC4647465
- DOI: 10.1261/rna.053397.115
Modeling of autosomal-dominant retinitis pigmentosa in Caenorhabditis elegans uncovers a nexus between global impaired functioning of certain splicing factors and cell type-specific apoptosis
Abstract
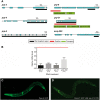
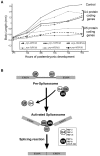
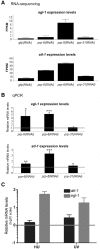
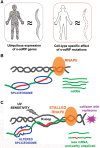
Retinitis pigmentosa (RP) is a rare genetic disease that causes gradual blindness through retinal degeneration. Intriguingly, seven of the 24 genes identified as responsible for the autosomal-dominant form (adRP) are ubiquitous spliceosome components whose impairment causes disease only in the retina. The fact that these proteins are essential in all organisms hampers genetic, genomic, and physiological studies, but we addressed these difficulties by using RNAi in Caenorhabditis elegans. Our study of worm phenotypes produced by RNAi of splicing-related adRP (s-adRP) genes functionally distinguishes between components of U4 and U5 snRNP complexes, because knockdown of U5 proteins produces a stronger phenotype. RNA-seq analyses of worms where s-adRP genes were partially inactivated by RNAi, revealed mild intron retention in developing animals but not in adults, suggesting a positive correlation between intron retention and transcriptional activity. Interestingly, RNAi of s-adRP genes produces an increase in the expression of atl-1 (homolog of human ATR), which is normally activated in response to replicative stress and certain DNA-damaging agents. The up-regulation of atl-1 correlates with the ectopic expression of the pro-apoptotic gene egl-1 and apoptosis in hypodermal cells, which produce the cuticle, but not in other cell types. Our model in C. elegans resembles s-adRP in two aspects: The phenotype caused by global knockdown of s-adRP genes is cell type-specific and associated with high transcriptional activity. Finally, along with a reduced production of mature transcripts, we propose a model in which the retina-specific cell death in s-adRP patients can be induced through genomic instability.
Keywords: C. elegans; RNA-seq; intron retention; retinitis pigmentosa; spliceosome.
© 2015 Rubio-Peña et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




Similar articles
-
Hypoxia-regulated components of the U4/U6.U5 tri-small nuclear riboprotein complex: possible role in autosomal dominant retinitis pigmentosa.Mol Vis. 2008 Jan 25;14:125-35. Mol Vis. 2008. PMID: 18334927 Free PMC article.
-
Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs.Am J Hum Genet. 2009 Nov;85(5):617-27. doi: 10.1016/j.ajhg.2009.09.020. Epub 2009 Oct 29. Am J Hum Genet. 2009. PMID: 19878916 Free PMC article.
-
Mimicking of splicing-related retinitis pigmentosa mutations in C. elegans allow drug screens and identification of disease modifiers.Hum Mol Genet. 2020 Mar 27;29(5):756-765. doi: 10.1093/hmg/ddz315. Hum Mol Genet. 2020. PMID: 31919495
-
Pre-mRNA splicing and retinitis pigmentosa.Mol Vis. 2006 Oct 26;12:1259-71. Mol Vis. 2006. PMID: 17110909 Free PMC article. Review.
-
[Progress in pathogenesis and therapeutic research in retinitis pigmentosa and age-related macular degeneration].Nippon Ganka Gakkai Zasshi. 2004 Dec;108(12):750-68; discussion 769. Nippon Ganka Gakkai Zasshi. 2004. PMID: 15656086 Review. Japanese.
Cited by
-
Caenorhabditis elegans for research on cancer hallmarks.Dis Model Mech. 2023 Jun 1;16(6):dmm050079. doi: 10.1242/dmm.050079. Epub 2023 Jun 6. Dis Model Mech. 2023. PMID: 37278614 Free PMC article.
-
mRNA Editing, Processing and Quality Control in Caenorhabditis elegans.Genetics. 2020 Jul;215(3):531-568. doi: 10.1534/genetics.119.301807. Genetics. 2020. PMID: 32632025 Free PMC article. Review.
-
Addressing the tissue specificity of U5 snRNP spliceosomopathies.Front Cell Dev Biol. 2025 Apr 8;13:1572188. doi: 10.3389/fcell.2025.1572188. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40264708 Free PMC article.
-
Mutations in the Spliceosome Component prp-6 and Overexpression of cdh-5 Suppress Axon Guidance Defects of cdh-4 Mutants in Caenorhabditis elegans.Neurosci Insights. 2022 Sep 6;17:26331055221123346. doi: 10.1177/26331055221123346. eCollection 2022. Neurosci Insights. 2022. PMID: 36090596 Free PMC article.
-
Application of CRISPR Tools for Variant Interpretation and Disease Modeling in Inherited Retinal Dystrophies.Genes (Basel). 2020 Apr 27;11(5):473. doi: 10.3390/genes11050473. Genes (Basel). 2020. PMID: 32349249 Free PMC article. Review.
References
-
- Abu-Safieh L, Vithana EN, Mantel I, Holder GE, Pelosini L, Bird AC, Bhattacharya SS. 2006. A large deletion in the adRP gene PRPF31: evidence that haploinsufficiency is the cause of disease. Mol Vis 12: 384–388. - PubMed
-
- Aguilera A, García-Muse T. 2012. R loops: from transcription byproducts to threats to genome stability. Mol Cell 46: 115–124. - PubMed
-
- Aguilera A, García-Muse T. 2013. Causes of genome instability. Annu Rev Genet 47: 1–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
