The spliceosome, a potential Achilles heel of MYC-driven tumors
- PMID: 26490253
- PMCID: PMC4618744
- DOI: 10.1186/s13073-015-0234-3
The spliceosome, a potential Achilles heel of MYC-driven tumors
Abstract
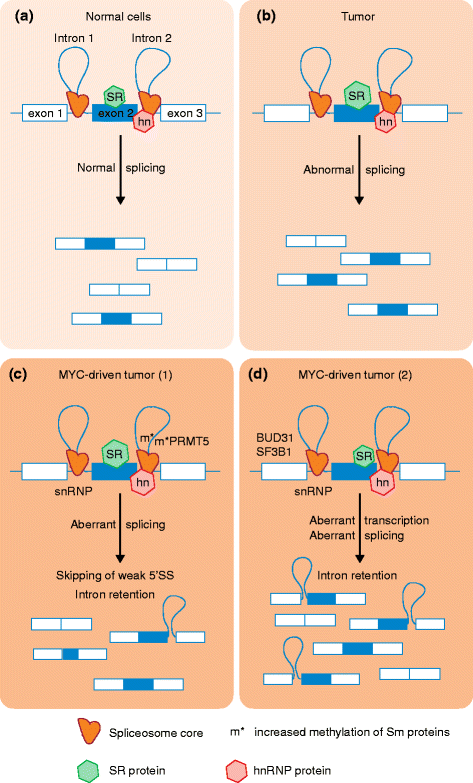
Alterations in RNA splicing are frequent in human tumors. Two recent studies of lymphoma and breast cancer have identified components of the spliceosome - the core splicing machinery - that are essential for malignant transformation driven by the transcription factor MYC. These findings provide a direct link between MYC and RNA splicing deregulation, and raise the exciting possibility of targeting spliceosome components in MYC-driven tumors.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

