The transcriptional landscape of age in human peripheral blood
- PMID: 26490707
- PMCID: PMC4639797
- DOI: 10.1038/ncomms9570
The transcriptional landscape of age in human peripheral blood
Abstract
Disease incidences increase with age, but the molecular characteristics of ageing that lead to increased disease susceptibility remain inadequately understood. Here we perform a whole-blood gene expression meta-analysis in 14,983 individuals of European ancestry (including replication) and identify 1,497 genes that are differentially expressed with chronological age. The age-associated genes do not harbor more age-associated CpG-methylation sites than other genes, but are instead enriched for the presence of potentially functional CpG-methylation sites in enhancer and insulator regions that associate with both chronological age and gene expression levels. We further used the gene expression profiles to calculate the 'transcriptomic age' of an individual, and show that differences between transcriptomic age and chronological age are associated with biological features linked to ageing, such as blood pressure, cholesterol levels, fasting glucose, and body mass index. The transcriptomic prediction model adds biological relevance and complements existing epigenetic prediction models, and can be used by others to calculate transcriptomic age in external cohorts.
Figures

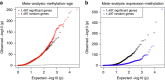
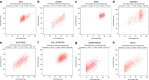
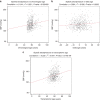
References
-
- Anselmi C. V. et al. Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 12, 95–104 (2009). - PubMed
-
- Nebel A. et al. A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech. Ageing Dev. 132, 324–330 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

