A Second β-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans
- PMID: 26491009
- PMCID: PMC4692217
- DOI: 10.1074/jbc.M115.688630
A Second β-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans
Abstract
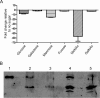
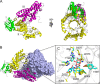
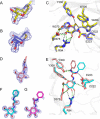
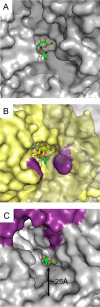
An important facet of the interaction between the pathogen Streptococcus pneumoniae (pneumococcus) and its human host is the ability of this bacterium to process host glycans. To achieve cleavage of the glycosidic bonds in host glycans, S. pneumoniae deploys a wide array of glycoside hydrolases. Here, we identify and characterize a new family 20 glycoside hydrolase, GH20C, from S. pneumoniae. Recombinant GH20C possessed the ability to hydrolyze the β-linkages joining either N-acetylglucosamine or N-acetylgalactosamine to a wide variety of aglycon residues, thus revealing this enzyme to be a generalist N-acetylhexosaminidase in vitro. X-ray crystal structures were determined for GH20C in a ligand-free form, in complex with the N-acetylglucosamine and N-acetylgalactosamine products of catalysis and in complex with both gluco- and galacto-configured inhibitors O-(2-acetamido-2-deoxy-D-glucopyranosylidene)amino N-phenyl carbamate (PUGNAc), O-(2-acetamido-2-deoxy-D-galactopyranosylidene)amino N-phenyl carbamate (GalPUGNAc), N-acetyl-D-glucosamine-thiazoline (NGT), and N-acetyl-D-galactosamine-thiazoline (GalNGT) at resolutions from 1.84 to 2.7 Å. These structures showed N-acetylglucosamine and N-acetylgalactosamine to be recognized via identical sets of molecular interactions. Although the same sets of interaction were maintained with the gluco- and galacto-configured inhibitors, the inhibition constants suggested preferred recognition of the axial O4 when an aglycon moiety was present (Ki for PUGNAc > GalPUGNAc) but preferred recognition of an equatorial O4 when the aglycon was absent (Ki for GalNGT > NGT). Overall, this study reveals GH20C to be another tool that is unique in the arsenal of S. pneumoniae and that it may implement the effort of the bacterium to utilize and/or destroy the wide array of host glycans that it may encounter.
Keywords: Streptococcus; enzyme inhibitor; glycobiology; glycoside hydrolase; x-ray crystallography.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


References
-
- Bøhle L. A., Mathiesen G., Vaaje-Kolstad G., and Eijsink V. G. (2011) An endo-β-N-acetylglucosaminidase from Enterococcus faecalis V583 responsible for the hydrolysis of high-mannose and hybrid-type N-linked glycans. FEMS Microbiol. Lett. 325, 123–129 - PubMed
-
- Prag G., Papanikolau Y., Tavlas G., Vorgias C. E., Petratos K., and Oppenheim A. B. (2000) Structures of chitobiase mutants complexed with the substrate di-N-acetyl-d-glucosamine: the catalytic role of the conserved acidic pair, aspartate 539 and glutamate 540. J. Mol. Biol. 300, 611–617 - PubMed
-
- Sumida T., Stubbs K. A., Ito M., and Yokoyama S. (2012) Gaining insight into the inhibition of glycoside hydrolase family 20 exo-β-N-acetylhexosaminidases using a structural approach. Org. Biomol. Chem. 10, 2607–2612 - PubMed
-
- Williams S. J., Mark B. L., Vocadlo D. J., James M. N., and Withers S. G. (2002) Aspartate 313 in the Streptomyces plicatus hexosaminidase plays a critical role in substrate-assisted catalysis by orienting the 2-acetamido group and stabilizing the transition state. J. Biol. Chem. 277, 40055–40065 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

