Functional Characterization of Glycoprotein H Chimeras Composed of Conserved Domains of the Pseudorabies Virus and Herpes Simplex Virus 1 Homologs
- PMID: 26491153
- PMCID: PMC4702552
- DOI: 10.1128/JVI.01985-15
Functional Characterization of Glycoprotein H Chimeras Composed of Conserved Domains of the Pseudorabies Virus and Herpes Simplex Virus 1 Homologs
Abstract
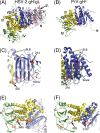
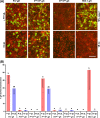
Membrane fusion is indispensable for entry of enveloped viruses into host cells. The conserved core fusion machinery of the Herpesviridae consists of glycoprotein B (gB) and the gH/gL complex. Recently, crystal structures of gH/gL of herpes simplex virus 2 (HSV-2) and Epstein-Barr virus and of a core fragment of pseudorabies virus (PrV) gH identified four structurally conserved gH domains. To investigate functional conservation, chimeric genes encoding combinations of individual domains of PrV and herpes simplex virus 1 (HSV-1) gH were expressed in rabbit kidney cells, and their processing and transport to the cell surface, as well as activity in fusion assays including gB, gD, and gL of PrV or HSV-1, were analyzed. Chimeric gH containing domain I of HSV-1 and domains II to IV of PrV exhibited limited fusion activity in the presence of PrV gB and gD and HSV-1 gL, but not of PrV gL. More strikingly, chimeric gH consisting of PrV domains I to III and HSV-1 domain IV exhibited considerable fusion activity together with PrV gB, gD, and gL. Replacing PrV gB with the HSV-1 protein significantly enhanced this activity. A cell line stably expressing this chimeric gH supported replication of gH-deleted PrV. Our results confirm the specificity of domain I for gL binding, demonstrate functional conservation of domain IV in two alphaherpesviruses from different genera, and indicate species-specific interactions of this domain with gB. They also suggest that gH domains II and III might form a structural and functional unit which does not tolerate major substitutions.
Importance: Envelope glycoprotein H (gH) is essential for herpesvirus-induced membrane fusion, which is required for host cell entry and viral spread. Although gH is structurally conserved within the Herpesviridae, its precise role and its interactions with other components of the viral fusion machinery are not fully understood. Chimeric proteins containing domains of gH proteins from different herpesviruses can serve as tools to elucidate the molecular basis of gH function. The present study shows that the C-terminal part of human herpesvirus 1 (herpes simplex virus 1) gH can functionally substitute for the corresponding part of suid herpesvirus 1 (pseudorabies virus) gH, whereas other tested combinations proved to be nonfunctional. Interestingly, the exchangeable fragment included the membrane-proximal end of the gH ectodomain (domain IV), which is most conserved in sequence and structure and might be capable of transient membrane interaction during fusion.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Functional Relevance of the Transmembrane Domain and Cytoplasmic Tail of the Pseudorabies Virus Glycoprotein H for Membrane Fusion.J Virol. 2018 May 29;92(12):e00376-18. doi: 10.1128/JVI.00376-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618646 Free PMC article.
-
Functional Relevance of the N-Terminal Domain of Pseudorabies Virus Envelope Glycoprotein H and Its Interaction with Glycoprotein L.J Virol. 2017 Apr 13;91(9):e00061-17. doi: 10.1128/JVI.00061-17. Print 2017 May 1. J Virol. 2017. PMID: 28228592 Free PMC article.
-
A Functional Interaction between Herpes Simplex Virus 1 Glycoprotein gH/gL Domains I and II and gD Is Defined by Using Alphaherpesvirus gH and gL Chimeras.J Virol. 2015 Jul;89(14):7159-69. doi: 10.1128/JVI.00740-15. Epub 2015 Apr 29. J Virol. 2015. PMID: 25926636 Free PMC article.
-
Two Sides to Every Story: Herpes Simplex Type-1 Viral Glycoproteins gB, gD, gH/gL, gK, and Cellular Receptors Function as Key Players in Membrane Fusion.Viruses. 2021 Sep 16;13(9):1849. doi: 10.3390/v13091849. Viruses. 2021. PMID: 34578430 Free PMC article. Review.
-
The structural basis of herpesvirus entry.Nat Rev Microbiol. 2021 Feb;19(2):110-121. doi: 10.1038/s41579-020-00448-w. Epub 2020 Oct 21. Nat Rev Microbiol. 2021. PMID: 33087881 Free PMC article. Review.
Cited by
-
Entry of Alphaherpesviruses.Curr Issues Mol Biol. 2021;41:63-124. doi: 10.21775/cimb.041.063. Epub 2020 Aug 7. Curr Issues Mol Biol. 2021. PMID: 32764159 Free PMC article. Review.
-
Influence of N-glycosylation on Expression and Function of Pseudorabies Virus Glycoprotein gB.Pathogens. 2021 Jan 12;10(1):61. doi: 10.3390/pathogens10010061. Pathogens. 2021. PMID: 33445487 Free PMC article.
-
Protein- and Peptide-Based Virus Inactivators: Inactivating Viruses Before Their Entry Into Cells.Front Microbiol. 2020 May 25;11:1063. doi: 10.3389/fmicb.2020.01063. eCollection 2020. Front Microbiol. 2020. PMID: 32523582 Free PMC article. Review.
-
Functional Relevance of the Transmembrane Domain and Cytoplasmic Tail of the Pseudorabies Virus Glycoprotein H for Membrane Fusion.J Virol. 2018 May 29;92(12):e00376-18. doi: 10.1128/JVI.00376-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29618646 Free PMC article.
-
Mechanism of herpesvirus UL24 protein regulating viral immune escape and virulence.Front Microbiol. 2023 Sep 22;14:1268429. doi: 10.3389/fmicb.2023.1268429. eCollection 2023. Front Microbiol. 2023. PMID: 37808279 Free PMC article. Review.
References
-
- Roizman B, Knipe DM, Whitney RJ. 2007. Herpes simplex viruses, p 2501–2601. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Atanasiu D, Saw WT, Gallagher JR, Hannah BP, Matsuda Z, Whitbeck JC, Cohen GH, Eisenberg RJ. 2013. Dual split protein-based fusion assay reveals that mutations to herpes simplex virus (HSV) glycoprotein gB alter the kinetics of cell-cell fusion induced by HSV entry glycoproteins. J Virol 87:11332–11345. doi: 10.1128/JVI.01700-13. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

