Identification of a Novel Viral Protein Expressed from the PB2 Segment of Influenza A Virus
- PMID: 26491155
- PMCID: PMC4702538
- DOI: 10.1128/JVI.02175-15
Identification of a Novel Viral Protein Expressed from the PB2 Segment of Influenza A Virus
Abstract
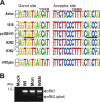
Over the past 2 decades, several novel influenza virus proteins have been identified that modulate viral infections in vitro and/or in vivo. The PB2 segment, which is one of the longest influenza A virus segments, is known to encode only one viral protein, PB2. In the present study, we used reverse transcription-PCR (RT-PCR) targeting viral mRNAs transcribed from the PB2 segment to look for novel viral proteins encoded by spliced mRNAs. We identified a new viral protein, PB2-S1, encoded by a novel spliced mRNA in which the region corresponding to nucleotides 1513 to 1894 of the PB2 mRNA is deleted. PB2-S1 was detected in virus-infected cells and in cells transfected with a protein expression plasmid encoding PB2. PB2-S1 localized to mitochondria, inhibited the RIG-I-dependent interferon signaling pathway, and interfered with viral polymerase activity (dependent on its PB1-binding capability). The nucleotide sequences around the splicing donor and acceptor sites for PB2-S1 were highly conserved among pre-2009 human H1N1 viruses but not among human H1N1pdm and H3N2 viruses. PB2-S1-deficient viruses, however, showed growth kinetics in MDCK cells and virulence in mice similar to those of wild-type virus. The biological significance of PB2-S1 to the replication and pathogenicity of seasonal H1N1 influenza A viruses warrants further investigation.
Importance: Transcriptome analysis of cells infected with influenza A virus has improved our understanding of the host response to viral infection, because such analysis yields considerable information about both in vitro and in vivo viral infections. However, little attention has been paid to transcriptomes derived from the viral genome. Here we focused on the splicing of mRNA expressed from the PB2 segment and identified a spliced viral mRNA encoding a novel viral protein. This result suggests that other, as yet unidentified viral proteins encoded by spliced mRNAs could be expressed in virus-infected cells. A viral transcriptome including the viral spliceosome should be evaluated to gain new insights into influenza virus infection.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures






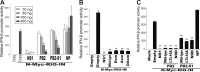


References
-
- Palese P, Ritchey MB, Schulman JL. 1977. Mapping of the influenza virus genome. II. Identification of the P1, P2, and P3 genes. Virology 76:114–121. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

