Structure/Function Studies Involving the V3 Region of the HIV-1 Envelope Delineate Multiple Factors That Affect Neutralization Sensitivity
- PMID: 26491157
- PMCID: PMC4702699
- DOI: 10.1128/JVI.01645-15
Structure/Function Studies Involving the V3 Region of the HIV-1 Envelope Delineate Multiple Factors That Affect Neutralization Sensitivity
Abstract
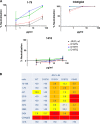
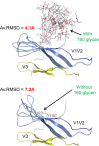
Antibodies (Abs) specific for the V3 loop of the HIV-1 gp120 envelope neutralize most tier 1 and many tier 2 viruses and are present in essentially all HIV-infected individuals as well as immunized humans and animals. Vaccine-induced V3 Abs are associated with reduced HIV infection rates in humans and affect the nature of transmitted viruses in infected vaccinees, despite the fact that V3 is often occluded in the envelope trimer. Here, we link structural and experimental data showing how conformational alterations of the envelope trimer render viruses exceptionally sensitive to V3 Abs. The experiments interrogated the neutralization sensitivity of pseudoviruses with single amino acid mutations in various regions of gp120 that were predicted to alter packing of the V3 loop in the Env trimer. The results indicate that the V3 loop is metastable in the envelope trimer on the virion surface, flickering between states in which V3 is either occluded or available for binding to chemokine receptors (leading to infection) and to V3 Abs (leading to virus neutralization). The spring-loaded V3 in the envelope trimer is easily released by disruption of the stability of the V3 pocket in the unliganded trimer or disruption of favorable V3/pocket interactions. Formation of the V3 pocket requires appropriate positioning of the V1V2 domain, which is, in turn, dependent on the conformation of the bridging sheet and on the stability of the V1V2 B-C strand-connecting loop.
Importance: The levels of antibodies to the third variable region (V3) of the HIV envelope protein correlate with reduced HIV infection rates. Previous studies showed that V3 is often occluded, as it sits in a pocket of the envelope trimer on the surface of virions; however, the trimer is flexible, allowing occluded portions of the envelope (like V3) to flicker into an exposed position that binds antibodies. Here we provide a systematic interrogation of mechanisms by which single amino acid changes in various regions of gp120 (i) render viruses sensitive to neutralization by V3 antibodies, (ii) result in altered packing of the V3 loop, and (iii) activate an open conformation that exposes V3 to the effects of V3 Abs. Taken together, these and previous studies explain how V3 antibodies can protect against HIV-1 infection and why they should be one of the targets of vaccine-induced antibodies.
Copyright © 2015 Zolla-Pazner et al.
Figures




Similar articles
-
Plasticity and Epitope Exposure of the HIV-1 Envelope Trimer.J Virol. 2017 Aug 10;91(17):e00410-17. doi: 10.1128/JVI.00410-17. Print 2017 Sep 1. J Virol. 2017. PMID: 28615206 Free PMC article.
-
Structural Constraints at the Trimer Apex Stabilize the HIV-1 Envelope in a Closed, Antibody-Protected Conformation.mBio. 2018 Dec 11;9(6):e00955-18. doi: 10.1128/mBio.00955-18. mBio. 2018. PMID: 30538178 Free PMC article.
-
Distinct mechanisms regulate exposure of neutralizing epitopes in the V2 and V3 loops of HIV-1 envelope.J Virol. 2014 Nov;88(21):12853-65. doi: 10.1128/JVI.02125-14. Epub 2014 Aug 27. J Virol. 2014. PMID: 25165106 Free PMC article.
-
Conformation-Dependent Interactions Between HIV-1 Envelope Glycoproteins and Broadly Neutralizing Antibodies.AIDS Res Hum Retroviruses. 2018 Sep;34(9):794-803. doi: 10.1089/AID.2018.0102. Epub 2018 Jul 17. AIDS Res Hum Retroviruses. 2018. PMID: 29905080 Review.
-
Improving on nature: focusing the immune response on the V3 loop.Hum Antibodies. 2005;14(3-4):69-72. Hum Antibodies. 2005. PMID: 16720976 Review.
Cited by
-
Functional and Highly Cross-Linkable HIV-1 Envelope Glycoproteins Enriched in a Pretriggered Conformation.J Virol. 2022 Apr 27;96(8):e0166821. doi: 10.1128/jvi.01668-21. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343783 Free PMC article.
-
Effects of the SOS (A501C/T605C) and DS (I201C/A433C) Disulfide Bonds on HIV-1 Membrane Envelope Glycoprotein Conformation and Function.J Virol. 2019 May 29;93(12):e00304-19. doi: 10.1128/JVI.00304-19. Print 2019 Jun 15. J Virol. 2019. PMID: 30944182 Free PMC article.
-
Broadly neutralizing antibody epitopes on HIV-1 particles are exposed after virus interaction with host cells.J Virol. 2023 Sep 28;97(9):e0071023. doi: 10.1128/jvi.00710-23. Epub 2023 Sep 8. J Virol. 2023. PMID: 37681958 Free PMC article.
-
Trapping the HIV-1 V3 loop in a helical conformation enables broad neutralization.Nat Struct Mol Biol. 2023 Sep;30(9):1323-1336. doi: 10.1038/s41594-023-01062-z. Epub 2023 Aug 21. Nat Struct Mol Biol. 2023. PMID: 37605043 Free PMC article.
-
VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.Cell Host Microbe. 2020 Jun 10;27(6):963-975.e5. doi: 10.1016/j.chom.2020.03.024. Epub 2020 Apr 20. Cell Host Microbe. 2020. PMID: 32315598 Free PMC article.
References
-
- Hill CM, Deng HK, Unutmaz D, KewalRamani VN, Bastiani L, Gorny MK, Zolla-Pazner S, Littman DR. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol 71:6296–6304. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

