Hepatocyte Heparan Sulfate Is Required for Adeno-Associated Virus 2 but Dispensable for Adenovirus 5 Liver Transduction In Vivo
- PMID: 26491162
- PMCID: PMC4702579
- DOI: 10.1128/JVI.01939-15
Hepatocyte Heparan Sulfate Is Required for Adeno-Associated Virus 2 but Dispensable for Adenovirus 5 Liver Transduction In Vivo
Abstract
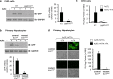
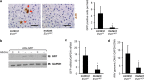
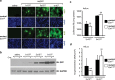
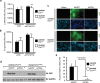
Adeno-associated virus 2 (AAV2) and adenovirus 5 (Ad5) are promising gene therapy vectors. Both display liver tropism and are currently thought to enter hepatocytes in vivo through cell surface heparan sulfate proteoglycans (HSPGs). To test directly this hypothesis, we created mice that lack Ext1, an enzyme required for heparan sulfate biosynthesis, in hepatocytes. Ext1(HEP) mutant mice exhibit an 8-fold reduction of heparan sulfate in primary hepatocytes and a 5-fold reduction of heparan sulfate in whole liver tissue. Conditional hepatocyte Ext1 gene deletion greatly reduced AAV2 liver transduction following intravenous injection. Ad5 transduction requires blood coagulation factor X (FX); FX binds to the Ad5 capsid hexon protein and bridges the virus to HSPGs on the cell surface. Ad5.FX transduction was abrogated in primary hepatocytes from Ext1(HEP) mice. However, in contrast to the case with AAV2, Ad5 transduction was not significantly reduced in the livers of Ext1(HEP) mice. FX remained essential for Ad5 transduction in vivo in Ext1(HEP) mice. We conclude that while AAV2 requires HSPGs for entry into mouse hepatocytes, HSPGs are dispensable for Ad5 hepatocyte transduction in vivo. This study reopens the question of how adenovirus enters cells in vivo.
Importance: Our understanding of how viruses enter cells, and how they can be used as therapeutic vectors to manage disease, begins with identification of the cell surface receptors to which viruses bind and which mediate viral entry. Both adeno-associated virus 2 and adenovirus 5 are currently thought to enter hepatocytes in vivo through heparan sulfate proteoglycans (HSPGs). However, direct evidence for these conclusions is lacking. Experiments presented herein, in which hepatic heparan sulfate synthesis was genetically abolished, demonstrated that HSPGs are not likely to function as hepatocyte Ad5 receptors in vivo. The data also demonstrate that HSPGs are required for hepatocyte transduction by AAV2. These results reopen the question of the identity of the Ad5 receptor in vivo and emphasize the necessity of demonstrating the nature of the receptor by genetic means, both for understanding Ad5 entry into cells in vivo and for optimization of Ad5 vectors as therapeutic agents.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures


Similar articles
-
Requirements for receptor engagement during infection by adenovirus complexed with blood coagulation factor X.PLoS Pathog. 2010 Oct 7;6(10):e1001142. doi: 10.1371/journal.ppat.1001142. PLoS Pathog. 2010. PMID: 20949078 Free PMC article.
-
Interaction between mouse adenovirus type 1 and cell surface heparan sulfate proteoglycans.PLoS One. 2012;7(2):e31454. doi: 10.1371/journal.pone.0031454. Epub 2012 Feb 7. PLoS One. 2012. PMID: 22347482 Free PMC article.
-
Differential effects of murine and human factor X on adenovirus transduction via cell-surface heparan sulfate.J Biol Chem. 2011 Jul 15;286(28):24535-43. doi: 10.1074/jbc.M111.241562. Epub 2011 May 19. J Biol Chem. 2011. PMID: 21596747 Free PMC article.
-
Tropism and transduction of oncolytic adenovirus 5 vectors in cancer therapy: Focus on fiber chimerism and mosaicism, hexon and pIX.Virus Res. 2018 Sep 15;257:40-51. doi: 10.1016/j.virusres.2018.08.012. Epub 2018 Aug 17. Virus Res. 2018. PMID: 30125593 Review.
-
Heparan sulphate proteoglycans and viral vectors : ally or foe?Curr Gene Ther. 2006 Feb;6(1):35-44. doi: 10.2174/156652306775515565. Curr Gene Ther. 2006. PMID: 16475944 Review.
Cited by
-
Blood Coagulation Factor X Exerts Differential Effects on Adenovirus Entry into Human Lymphocytes.Viruses. 2018 Jan 3;10(1):20. doi: 10.3390/v10010020. Viruses. 2018. PMID: 29301346 Free PMC article.
-
Hexons from adenovirus serotypes 5 and 48 differentially protect adenovirus vectors from neutralization by mouse and human serum.PLoS One. 2018 Feb 5;13(2):e0192353. doi: 10.1371/journal.pone.0192353. eCollection 2018. PLoS One. 2018. PMID: 29401488 Free PMC article.
-
PTEN knockdown with the Y444F mutant AAV2 vector promotes axonal regeneration in the adult optic nerve.Neural Regen Res. 2018 Jan;13(1):135-144. doi: 10.4103/1673-5374.224381. Neural Regen Res. 2018. PMID: 29451218 Free PMC article.
-
The relevance of coagulation factor X protection of adenoviruses in human sera.Gene Ther. 2016 Jul;23(7):592-6. doi: 10.1038/gt.2016.32. Epub 2016 Mar 25. Gene Ther. 2016. PMID: 27014840 Free PMC article.
-
Re-administration of AAV vectors by masking with host albumin: A Goldilocks hypothesis.Mol Ther. 2023 Jul 5;31(7):1870-1873. doi: 10.1016/j.ymthe.2023.06.009. Epub 2023 Jun 26. Mol Ther. 2023. PMID: 37369207 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

