COordination of Standards in MetabOlomicS (COSMOS): facilitating integrated metabolomics data access
- PMID: 26491418
- PMCID: PMC4605977
- DOI: 10.1007/s11306-015-0810-y
COordination of Standards in MetabOlomicS (COSMOS): facilitating integrated metabolomics data access
Abstract
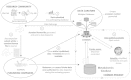
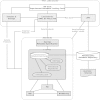
Metabolomics has become a crucial phenotyping technique in a range of research fields including medicine, the life sciences, biotechnology and the environmental sciences. This necessitates the transfer of experimental information between research groups, as well as potentially to publishers and funders. After the initial efforts of the metabolomics standards initiative, minimum reporting standards were proposed which included the concepts for metabolomics databases. Built by the community, standards and infrastructure for metabolomics are still needed to allow storage, exchange, comparison and re-utilization of metabolomics data. The Framework Programme 7 EU Initiative 'coordination of standards in metabolomics' (COSMOS) is developing a robust data infrastructure and exchange standards for metabolomics data and metadata. This is to support workflows for a broad range of metabolomics applications within the European metabolomics community and the wider metabolomics and biomedical communities' participation. Here we announce our concepts and efforts asking for re-engagement of the metabolomics community, academics and industry, journal publishers, software and hardware vendors, as well as those interested in standardisation worldwide (addressing missing metabolomics ontologies, complex-metadata capturing and XML based open source data exchange format), to join and work towards updating and implementing metabolomics standards.
Keywords: Coordination and data sharing community; Data exchange; Data standards; Metabolomics; Metabonomics; e-Infrastructure.
Figures
References
-
- Bais P, Moon SM, He K, Leitao R, Dreher K, Walk T, et al. PlantMetabolomics.org: a web portal for plant metabolomics experiments. [Research Support, U.S. Gov’t, Non-P.H.S.] Plant Physiology. 2010;152(4):1807–1816. doi: 10.1104/pp.109.151027. - DOI - PMC - PubMed
-
- Bernini P, Bertini I, Luchinat C, Nincheri P, Staderini S, Turano P. Standard operating procedures for pre-analytical handling of blood and urine for metabolomic studies and biobanks. [Research Support, Non-U.S. Gov’t] Journal of Biomolecular NMR. 2011;49(3–4):231–243. doi: 10.1007/s10858-011-9489-1. - DOI - PubMed
-
- Bundy J, Davey M, Viant M. Environmental metabolomics: A critical review and future perspectives. Metabolomics. 2009;5(1):3–21. doi: 10.1007/s11306-008-0152-0. - DOI
-
- Castle AL, Fiehn O, Kaddurah-Daouk R, Lindon JC. Metabolomics standards workshop and the development of international standards for reporting metabolomics experimental results. [Consensus Development Conference, NIH] Briefings in Bioinformatics. 2006;7(2):159–165. doi: 10.1093/bib/bbl008. - DOI - PubMed
Grants and funding
- BB/D006422/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I000933/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/C519038/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/I000771/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MC_UP_A090_1006/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
