DNA Methylation Landscapes of Human Fetal Development
- PMID: 26492326
- PMCID: PMC4619663
- DOI: 10.1371/journal.pgen.1005583
DNA Methylation Landscapes of Human Fetal Development
Abstract
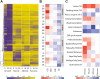
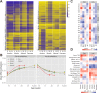
Remodelling the methylome is a hallmark of mammalian development and cell differentiation. However, current knowledge of DNA methylation dynamics in human tissue specification and organ development largely stems from the extrapolation of studies in vitro and animal models. Here, we report on the DNA methylation landscape using the 450k array of four human tissues (amnion, muscle, adrenal and pancreas) during the first and second trimester of gestation (9,18 and 22 weeks). We show that a tissue-specific signature, constituted by tissue-specific hypomethylated CpG sites, was already present at 9 weeks of gestation (W9). Furthermore, we report large-scale remodelling of DNA methylation from W9 to W22. Gain of DNA methylation preferentially occurred near genes involved in general developmental processes, whereas loss of DNA methylation mapped to genes with tissue-specific functions. Dynamic DNA methylation was associated with enhancers, but not promoters. Comparison of our data with external fetal adrenal, brain and liver revealed striking similarities in the trajectory of DNA methylation during fetal development. The analysis of gene expression data indicated that dynamic DNA methylation was associated with the progressive repression of developmental programs and the activation of genes involved in tissue-specific processes. The DNA methylation landscape of human fetal development provides insight into regulatory elements that guide tissue specification and lead to organ functionality.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Hon GC, Rajagopal N, Shen Y, McCleary DF, Yue F, Dang MD, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved.; 2013;45: 1198–206. 10.1038/ng.2746 - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

