GREM1 and POLE variants in hereditary colorectal cancer syndromes
- PMID: 26493165
- PMCID: PMC5057327
- DOI: 10.1002/gcc.22314
GREM1 and POLE variants in hereditary colorectal cancer syndromes
Abstract
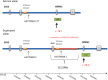
Hereditary factors are thought to play a role in at least one third of patients with colorectal cancer (CRC) but only a limited proportion of these have mutations in known high-penetrant genes. In a relatively large part of patients with a few or multiple colorectal polyps the underlying genetic cause of the disease is still unknown. Using exome sequencing in combination with linkage analyses together with detection of copy-number variations (CNV), we have identified a duplication in the regulatory region of the GREM1 gene in a family with an attenuated/atypical polyposis syndrome. In addition, 107 patients with colorectal cancer and/or polyposis were analyzed for mutations in the candidate genes identified. We also performed screening of the exonuclease domain of the POLE gene in a subset of these patients. The duplication of 16 kb in the regulatory region of GREM1 was found to be disease-causing in the family. Functional analyses revealed a higher expression of the GREM1 gene in colorectal tissue in duplication carriers. Screening of the exonuclease domain of POLE in additional CRC patients identified a probable causative novel variant c.1274A>G, p.Lys425Arg. In conclusion a high penetrant duplication in the regulatory region of GREM1, predisposing to CRC, was identified in a family with attenuated/atypical polyposis. A POLE variant was identified in a patient with early onset CRC and a microsatellite stable (MSS) tumor. Mutations leading to increased expression of genes can constitute disease-causing mutations in hereditary CRC syndromes.
© 2015 The Authors. Genes, Chromosomes & Cancer Published by Wiley Periodicals, Inc.
Figures




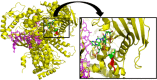
References
-
- Aaltonen L, Johns L, Jarvinen H, Mecklin JP, Houlston R. 2007. Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)‐deficient and MMR‐stable tumors. Clin Cancer Res 13:356–361. - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel‐Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

