Bcl-2 phosphorylation confers resistance on chronic lymphocytic leukaemia cells to the BH3 mimetics ABT-737, ABT-263 and ABT-199 by impeding direct binding
- PMID: 26493374
- PMCID: PMC4728412
- DOI: 10.1111/bph.13370
Bcl-2 phosphorylation confers resistance on chronic lymphocytic leukaemia cells to the BH3 mimetics ABT-737, ABT-263 and ABT-199 by impeding direct binding
Abstract
Background and purpose: Although the ongoing clinical trials of ABT-263 and ABT-199 in chronic lymphocytic leukaemia (CLL) have indicated that BH3 mimetics hold considerable promise, understanding the mechanism of CLL resistance to BH3 mimetics remains a challenge.
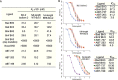
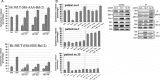
Experimental approach: The LD50 values of ABT-737, ABT-263 and ABT-199 in a number of primary CLL cells from 40 patients, were determined. The levels of Bcl-2 family proteins, including phosphorylated Bcl-2 (pBcl-2) and their interactions were measured by immunoblotting and co-immunoprecipitation. In vitro binding assays were performed by isothermal titration calorimetry and ELISA. BH3 profiling in isolated mitochondria was analysed.
Key results: The ratio of (Mcl-1 + pBcl-2) to Bcl-2 expression provided the most significant predictive marker for the cytotoxic potential of ABT-737, ABT-263 and ABT-199 in the panel of CLL samples. Mechanistically, pBcl-2 inhibited the effects of the ABT compounds on the displacement of Bax and Bim from Bcl-2, thereby suppressing mitochondrial apoptosis. The ABT compounds exhibited 100-300-fold lower binding affinity to the glutamic acid, phosphomimetic, mutant of Bcl-2 (T69E, S70E and S87E; EEE-Bcl-2). BH3 peptides exhibited different rank orders of binding affinities to full-length WT-Bcl-2 and full-length EEE-Bcl-2.
Conclusions and implications: Our study suggested that a structural alteration in the BH3-binding groove was induced by phosphorylation of Bcl-2. Our data also provided a framework to overcome resistance of CLL cells to the ABT compounds by combining pBcl-2 kinase inhibitors with the ABT compounds.
© 2015 The British Pharmacological Society.
Figures


Similar articles
-
Cyclin E/Cdk2-dependent phosphorylation of Mcl-1 determines its stability and cellular sensitivity to BH3 mimetics.Oncotarget. 2015 Jul 10;6(19):16912-25. doi: 10.18632/oncotarget.4857. Oncotarget. 2015. PMID: 26219338 Free PMC article.
-
Impact of elevated anti-apoptotic MCL-1 and BCL-2 on the development and treatment of MLL-AF9 AML in mice.Cell Death Differ. 2019 Jul;26(7):1316-1331. doi: 10.1038/s41418-018-0209-1. Epub 2018 Nov 23. Cell Death Differ. 2019. PMID: 30470795 Free PMC article.
-
Selective BCL-2 inhibition by ABT-199 causes on-target cell death in acute myeloid leukemia.Cancer Discov. 2014 Mar;4(3):362-75. doi: 10.1158/2159-8290.CD-13-0609. Epub 2013 Dec 17. Cancer Discov. 2014. PMID: 24346116 Free PMC article.
-
The first MCL-1-selective BH3 mimetics have therapeutic potential for chronic lymphocytic leukemia.Crit Rev Oncol Hematol. 2016 Apr;100:32-6. doi: 10.1016/j.critrevonc.2016.02.003. Epub 2016 Feb 11. Crit Rev Oncol Hematol. 2016. PMID: 26899021 Review.
-
BH3 mimetics: status of the field and new developments.Mol Cancer Ther. 2013 Sep;12(9):1691-700. doi: 10.1158/1535-7163.MCT-13-0058. Epub 2013 Aug 23. Mol Cancer Ther. 2013. PMID: 23974697 Review.
Cited by
-
Mitochondrial PKM2 regulates oxidative stress-induced apoptosis by stabilizing Bcl2.Cell Res. 2017 Mar;27(3):329-351. doi: 10.1038/cr.2016.159. Epub 2016 Dec 30. Cell Res. 2017. PMID: 28035139 Free PMC article.
-
Targeting BCL-2 in B-cell malignancies and overcoming therapeutic resistance.Cell Death Dis. 2020 Nov 2;11(11):941. doi: 10.1038/s41419-020-03144-y. Cell Death Dis. 2020. PMID: 33139702 Free PMC article. Review.
-
Prime, shock and kill: BCL-2 inhibition for HIV cure.Front Immunol. 2022 Oct 20;13:1033609. doi: 10.3389/fimmu.2022.1033609. eCollection 2022. Front Immunol. 2022. PMID: 36341439 Free PMC article. Review.
-
Targeting the IKZF1/BCL-2 axis as a novel therapeutic strategy for treating acute T-cell lymphoblastic leukemia.Cancer Biol Ther. 2025 Dec;26(1):2457777. doi: 10.1080/15384047.2025.2457777. Epub 2025 Jan 25. Cancer Biol Ther. 2025. PMID: 39862423 Free PMC article.
-
Combination of bortezomib and venetoclax targets the pro-survival function of LMP-1 and EBNA-3C of Epstein-Barr virus in spontaneous lymphoblastoid cell lines.PLoS Pathog. 2024 Sep 26;20(9):e1012250. doi: 10.1371/journal.ppat.1012250. eCollection 2024 Sep. PLoS Pathog. 2024. PMID: 39325843 Free PMC article.
References
-
- Basu A, Haldar S (1998). Microtubule‐damaging drugs triggered bcl2 phosphorylation‐requirement of phosphorylation on both serine‐70 and serine‐87 residues of bcl2 protein. Int J Oncol 13: 659–664. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

